ChIP bias as a function of cross-linking time
- PMID: 26685864
- PMCID: PMC4860130
- DOI: 10.1007/s10577-015-9509-1
ChIP bias as a function of cross-linking time
Abstract
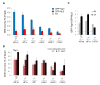
The chromatin immunoprecipitation (ChIP) assay is widely used to capture interactions between chromatin and regulatory proteins in vivo. Formaldehyde cross-linking of DNA and proteins is a critical step required to trap their interactions inside the cells before immunoprecipitation and analysis. Yet insufficient attention has been given to variables that might give rise to artifacts in this procedure, such as the duration of cross-linking. We analyzed the dependence of the ChIP signal on the duration of formaldehyde cross-linking time for two proteins: DNA topoisomerase 1 (Top1) that is functionally associated with the double helix in vivo, especially with active chromatin, and green fluorescent protein (GFP) that has no known bona fide interactions with DNA. With short time of formaldehyde fixation, only Top1 immunoprecipation efficiently recovered DNA from active promoters, whereas prolonged fixation augmented non-specific recovery of GFP dramatizing the need to optimize ChIP protocols to minimize the time of cross-linking, especially for abundant nuclear proteins. Thus, ChIP is a powerful approach to study the localization of protein on the genome when care is taken to manage potential artifacts.
Keywords: Chromatin immunoprecipitation; DNA topoisomerase 1; Formaldehyde cross-linking; Green fluorescent protein.
Figures



References
-
- Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53(6):937–47. - PubMed
-
- Gilmour DS, et al. Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell. 1986;44(3):401–7. - PubMed
-
- Gueron M, Kochoyan M, Leroy JL. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987;328(6125):89–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

