Reverse Engineering and Evaluation of Prediction Models for Progression to Type 2 Diabetes: An Application of Machine Learning Using Electronic Health Records
- PMID: 26685993
- PMCID: PMC4738229
- DOI: 10.1177/1932296815620200
Reverse Engineering and Evaluation of Prediction Models for Progression to Type 2 Diabetes: An Application of Machine Learning Using Electronic Health Records
Abstract
Background: Application of novel machine learning approaches to electronic health record (EHR) data could provide valuable insights into disease processes. We utilized this approach to build predictive models for progression to prediabetes and type 2 diabetes (T2D).
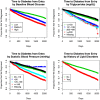
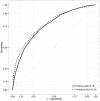
Methods: Using a novel analytical platform (Reverse Engineering and Forward Simulation [REFS]), we built prediction model ensembles for progression to prediabetes or T2D from an aggregated EHR data sample. REFS relies on a Bayesian scoring algorithm to explore a wide model space, and outputs a distribution of risk estimates from an ensemble of prediction models. We retrospectively followed 24 331 adults for transitions to prediabetes or T2D, 2007-2012. Accuracy of prediction models was assessed using an area under the curve (AUC) statistic, and validated in an independent data set.
Results: Our primary ensemble of models accurately predicted progression to T2D (AUC = 0.76), and was validated out of sample (AUC = 0.78). Models of progression to T2D consisted primarily of established risk factors (blood glucose, blood pressure, triglycerides, hypertension, lipid disorders, socioeconomic factors), whereas models of progression to prediabetes included novel factors (high-density lipoprotein, alanine aminotransferase, C-reactive protein, body temperature; AUC = 0.70).
Conclusions: We constructed accurate prediction models from EHR data using a hypothesis-free machine learning approach. Identification of established risk factors for T2D serves as proof of concept for this analytical approach, while novel factors selected by REFS represent emerging areas of T2D research. This methodology has potentially valuable downstream applications to personalized medicine and clinical research.
Keywords: diabetes mellitus; disease progression; electronic health records; medical informatics; prediabetic state; type 2.
© 2015 Diabetes Technology Society.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

