Actinomycin D Specifically Reduces Expanded CUG Repeat RNA in Myotonic Dystrophy Models
- PMID: 26686629
- PMCID: PMC4691565
- DOI: 10.1016/j.celrep.2015.11.028
Actinomycin D Specifically Reduces Expanded CUG Repeat RNA in Myotonic Dystrophy Models
Abstract
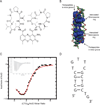
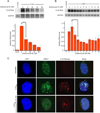
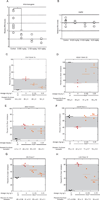
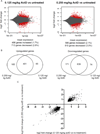
Myotonic dystrophy type 1 (DM1) is an inherited disease characterized by the inability to relax contracted muscles. Affected individuals carry large CTG expansions that are toxic when transcribed. One possible treatment approach is to reduce or eliminate transcription of CTG repeats. Actinomycin D (ActD) is a potent transcription inhibitor and FDA-approved chemotherapeutic that binds GC-rich DNA with high affinity. Here, we report that ActD decreased CUG transcript levels in a dose-dependent manner in DM1 cell and mouse models at significantly lower concentrations (nanomolar) compared to its use as a general transcription inhibitor or chemotherapeutic. ActD also significantly reversed DM1-associated splicing defects in a DM1 mouse model, and did so within the currently approved human treatment range. RNA-seq analyses showed that low concentrations of ActD did not globally inhibit transcription in a DM1 mouse model. These results indicate that transcription inhibition of CTG expansions is a promising treatment approach for DM1.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Chaouch S, Mouly V, Goyenvalle A, Vulin A, Mamchaoui K, Negroni E, Di Santo J, Butler-Browne G, Torrente Y, Garcia L, et al. Immortalized skin fibroblasts expressing conditional MyoD as a renewable and reliable source of converted human muscle cells to assess therapeutic strategies for muscular dystrophies: validation of an exon-skipping approach to restore dystrophin in Duchenne muscular dystrophy cells. Hum Gene Ther. 2009;20:784–790. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

