Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling
- PMID: 26686653
- PMCID: PMC4783382
- DOI: 10.1016/j.cell.2015.11.045
Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling
Abstract
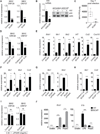
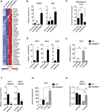
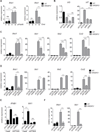
Cellular lipid requirements are achieved through a combination of biosynthesis and import programs. Using isotope tracer analysis, we show that type I interferon (IFN) signaling shifts the balance of these programs by decreasing synthesis and increasing import of cholesterol and long chain fatty acids. Genetically enforcing this metabolic shift in macrophages is sufficient to render mice resistant to viral challenge, demonstrating the importance of reprogramming the balance of these two metabolic pathways in vivo. Unexpectedly, mechanistic studies reveal that limiting flux through the cholesterol biosynthetic pathway spontaneously engages a type I IFN response in a STING-dependent manner. The upregulation of type I IFNs was traced to a decrease in the pool size of synthesized cholesterol and could be inhibited by replenishing cells with free cholesterol. Taken together, these studies delineate a metabolic-inflammatory circuit that links perturbations in cholesterol biosynthesis with activation of innate immunity.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



Comment in
-
How Low Cholesterol Is Good for Anti-viral Immunity.Cell. 2015 Dec 17;163(7):1572-4. doi: 10.1016/j.cell.2015.12.004. Cell. 2015. PMID: 26687349
-
Immunometabolism: The antiviral effect of limiting lipids.Nat Rev Immunol. 2016 Feb;16(2):76-7. doi: 10.1038/nri.2016.6. Epub 2016 Jan 19. Nat Rev Immunol. 2016. PMID: 26781936 No abstract available.
References
-
- Bard M, Sturm AM, Pierson CA, Brown S, Rogers KM, Nabinger S, Eckstein J, Barbuch R, Lees ND, Howell SA, et al. Sterol uptake in Candida glabrata: rescue of sterol auxotrophic strains. Diagnostic microbiology and infectious disease. 2005;52:285–293. - PubMed
-
- Castrillo A, Joseph SB, Vaidya SA, Haberland M, Fogelman AM, Cheng G, Tontonoz P. Crosstalk between LXR and toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism. Molecular cell. 2003;12:805–816. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- AI093768/AI/NIAID NIH HHS/United States
- UL1TR000124/TR/NCATS NIH HHS/United States
- R01 CA177322/CA/NCI NIH HHS/United States
- R21 HL126556/HL/NHLBI NIH HHS/United States
- T32 GM008042/GM/NIGMS NIH HHS/United States
- R01 AI084914/AI/NIAID NIH HHS/United States
- R21 DE023519/DE/NIDCR NIH HHS/United States
- R01 AI085043/AI/NIAID NIH HHS/United States
- S10 RR026744/RR/NCRR NIH HHS/United States
- P01 HL090553/HL/NHLBI NIH HHS/United States
- S10RR026744/RR/NCRR NIH HHS/United States
- P01 HL028481/HL/NHLBI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- T32 AI 060567/AI/NIAID NIH HHS/United States
- P30 AI028697/AI/NIAID NIH HHS/United States
- R21 AI110306/AI/NIAID NIH HHS/United States
- T32 AI060567/AI/NIAID NIH HHS/United States
- P01HL028481/HL/NHLBI NIH HHS/United States
- R01 AI093768/AI/NIAID NIH HHS/United States
- T32 GM008469/GM/NIGMS NIH HHS/United States
- R00 HL118161/HL/NHLBI NIH HHS/United States
- P01 CA177322/CA/NCI NIH HHS/United States
- HL126556/HL/NHLBI NIH HHS/United States
- R01 CA091791/CA/NCI NIH HHS/United States
- R01 DE023519/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

