Many lncRNAs, 5'UTRs, and pseudogenes are translated and some are likely to express functional proteins
- PMID: 26687005
- PMCID: PMC4739776
- DOI: 10.7554/eLife.08890
Many lncRNAs, 5'UTRs, and pseudogenes are translated and some are likely to express functional proteins
Abstract
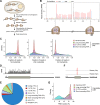
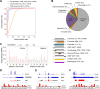
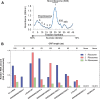
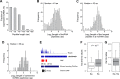
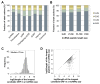
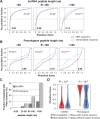
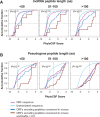
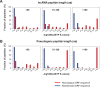
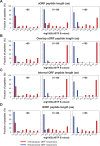
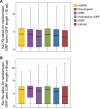
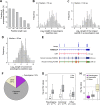
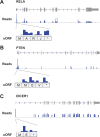
Using a new bioinformatic method to analyze ribosome profiling data, we show that 40% of lncRNAs and pseudogene RNAs expressed in human cells are translated. In addition, ~35% of mRNA coding genes are translated upstream of the primary protein-coding region (uORFs) and 4% are translated downstream (dORFs). Translated lncRNAs preferentially localize in the cytoplasm, whereas untranslated lncRNAs preferentially localize in the nucleus. The translation efficiency of cytoplasmic lncRNAs is nearly comparable to that of mRNAs, suggesting that cytoplasmic lncRNAs are engaged by the ribosome and translated. While most peptides generated from lncRNAs may be highly unstable byproducts without function, ~9% of the peptides are conserved in ORFs in mouse transcripts, as are 74% of pseudogene peptides, 24% of uORF peptides and 32% of dORF peptides. Analyses of synonymous and nonsynonymous substitution rates of these conserved peptides show that some are under stabilizing selection, suggesting potential functional importance.
Keywords: 5'UTR; biological function; cell biology; evolutionary biology; genomics; human; non-coding RNAs; pseudogene; ribosome profiling; translation.
Conflict of interest statement
KS: Reviewing editor,
The other authors declare that no competing interests exist.
Figures







Comment in
-
Translation and protein expression of lncRNAs: Impact for liver disease and hepatocellular carcinoma.Hepatology. 2016 Aug;64(2):671-4. doi: 10.1002/hep.28653. Epub 2016 Jun 24. Hepatology. 2016. PMID: 27228461 Free PMC article. No abstract available.
References
-
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. The EMBO Journal. 2014;33:981–993. doi: 10.1002/embj.201488411. - DOI - PMC - PubMed
-
- Bosley KS, Botchan M, Bredenoord AL, Carroll D, Charo RA, Charpentier E, Cohen R, Corn J, Doudna J, Feng G, Greely HT, Isasi R, Ji W, Kim J-S, Knoppers B, Lanphier E, Li J, Lovell-Badge R, Martin GS, Moreno J, Naldini L, Pera M, Perry ACF, Venter JC, Zhang F, Zhou Q, Regev A, Struhl K. CRISPR germline engineering—the community speaks. Nature Biotechnology. 2015;33:478–486. doi: 10.1038/nbt.3227. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

