Shadow Enhancers Are Pervasive Features of Developmental Regulatory Networks
- PMID: 26687625
- PMCID: PMC4712172
- DOI: 10.1016/j.cub.2015.11.034
Shadow Enhancers Are Pervasive Features of Developmental Regulatory Networks
Abstract
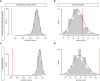
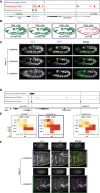
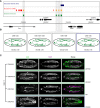
Embryogenesis is remarkably robust to segregating mutations and environmental variation; under a range of conditions, embryos of a given species develop into stereotypically patterned organisms. Such robustness is thought to be conferred, in part, through elements within regulatory networks that perform similar, redundant tasks. Redundant enhancers (or "shadow" enhancers), for example, can confer precision and robustness to gene expression, at least at individual, well-studied loci. However, the extent to which enhancer redundancy exists and can thereby have a major impact on developmental robustness remains unknown. Here, we systematically assessed this, identifying over 1,000 predicted shadow enhancers during Drosophila mesoderm development. The activity of 23 elements, associated with five genes, was examined in transgenic embryos, while natural structural variation among individuals was used to assess their ability to buffer against genetic variation. Our results reveal three clear properties of enhancer redundancy within developmental systems. First, it is much more pervasive than previously anticipated, with 64% of loci examined having shadow enhancers. Their spatial redundancy is often partial in nature, while the non-overlapping function may explain why these enhancers are maintained within a population. Second, over 70% of loci do not follow the simple situation of having only two shadow enhancers-often there are three (rols), four (CadN and ade5), or five (Traf1), at least one of which can be deleted with no obvious phenotypic effects. Third, although shadow enhancers can buffer variation, patterns of segregating variation suggest that they play a more complex role in development than generally considered.
Keywords: development; enhancer; redundancy; robustness; shadow enhancer; transcriptional networks.
Copyright © 2016 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures




References
-
- Montavon T., Soshnikova N., Mascrez B., Joye E., Thevenet L., Splinter E., de Laat W., Spitz F., Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

