A Multiplexed System for Quantitative Comparisons of Chromatin Landscapes
- PMID: 26687680
- PMCID: PMC4707994
- DOI: 10.1016/j.molcel.2015.11.003
A Multiplexed System for Quantitative Comparisons of Chromatin Landscapes
Abstract
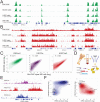
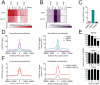
Genome-wide profiling of histone modifications can provide systematic insight into the regulatory elements and programs engaged in a given cell type. However, conventional chromatin immunoprecipitation and sequencing (ChIP-seq) does not capture quantitative information on histone modification levels, requires large amounts of starting material, and involves tedious processing of each individual sample. Here, we address these limitations with a technology that leverages DNA barcoding to profile chromatin quantitatively and in multiplexed format. We concurrently map relative levels of multiple histone modifications across multiple samples, each comprising as few as a thousand cells. We demonstrate the technology by monitoring dynamic changes following inhibition of p300, EZH2, or KDM5, by linking altered epigenetic landscapes to chromatin regulator mutations, and by mapping active and repressive marks in purified human hematopoietic stem cells. Hence, this technology enables quantitative studies of chromatin state dynamics across rare cell types, genotypes, environmental conditions, and drug treatments.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


References
-
- Brind’Amour J, Liu S, Hudson M, Chen C, Karimi M, Lorincz M. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Comms. 2015:6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

