INO1 transcriptional memory leads to DNA zip code-dependent interchromosomal clustering
- PMID: 26688804
- PMCID: PMC4682904
- DOI: 10.15698/mic2015.12.242
INO1 transcriptional memory leads to DNA zip code-dependent interchromosomal clustering
Abstract
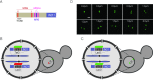
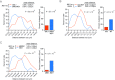
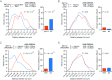
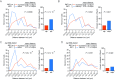
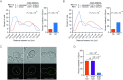
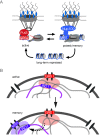
Many genes localize at the nuclear periphery through physical interaction with the nuclear pore complex (NPC). We have found that the yeast INO1 gene is targeted to the NPC both upon activation and for several generations after repression, a phenomenon called epigenetic transcriptional memory. Targeting of INO1 to the NPC requires distinct cis-acting promoter DNA zip codes under activating conditions and under memory conditions. When at the nuclear periphery, active INO1 clusters with itself and with other genes that share the GRS I zip code. Here, we show that during memory, the two alleles of INO1 cluster in diploids and endogenous INO1 clusters with an ectopic INO1 in haploids. After repression, INO1 does not cluster with GRS I - containing genes. Furthermore, clustering during memory requires Nup100 and two sets of DNA zip codes, those that target INO1 to the periphery when active and those that target it to the periphery after repression. Therefore, the interchromosomal clustering of INO1 that occurs during transcriptional memory is dependent upon, but mechanistically distinct from, the clustering of active INO1. Finally, while localization to the nuclear periphery is not regulated through the cell cycle during memory, clustering of INO1 during memory is regulated through the cell cycle.
Keywords: DNA zip code; epigenetic inheritance; interchromosomal clustering; nuclear pore; transcriptional memory.
Conflict of interest statement
Conflict of interest: The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
