Engineering an allosteric transcription factor to respond to new ligands
- PMID: 26689263
- PMCID: PMC4907361
- DOI: 10.1038/nmeth.3696
Engineering an allosteric transcription factor to respond to new ligands
Abstract
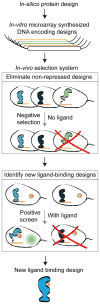
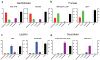
Genetic regulatory proteins inducible by small molecules are useful synthetic biology tools as sensors and switches. Bacterial allosteric transcription factors (aTFs) are a major class of regulatory proteins, but few aTFs have been redesigned to respond to new effectors beyond natural aTF-inducer pairs. Altering inducer specificity in these proteins is difficult because substitutions that affect inducer binding may also disrupt allostery. We engineered an aTF, the Escherichia coli lac repressor, LacI, to respond to one of four new inducer molecules: fucose, gentiobiose, lactitol and sucralose. Using computational protein design, single-residue saturation mutagenesis or random mutagenesis, along with multiplex assembly, we identified new variants comparable in specificity and induction to wild-type LacI with its inducer, isopropyl β-D-1-thiogalactopyranoside (IPTG). The ability to create designer aTFs will enable applications including dynamic control of cell metabolism, cell biology and synthetic gene circuits.
Figures



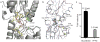
Similar articles
-
Altering residues N125 and D149 impacts sugar effector binding and allosteric parameters in Escherichia coli lactose repressor.Biochemistry. 2011 Oct 25;50(42):9002-13. doi: 10.1021/bi200896t. Epub 2011 Sep 30. Biochemistry. 2011. PMID: 21928765 Free PMC article.
-
A single mutation in the core domain of the lac repressor reduces leakiness.Microb Cell Fact. 2013 Jul 8;12:67. doi: 10.1186/1475-2859-12-67. Microb Cell Fact. 2013. PMID: 23834731 Free PMC article.
-
A Biosensor Strategy for E. coli Based on Ligand-Dependent Stabilization.ACS Synth Biol. 2018 Sep 21;7(9):1990-1999. doi: 10.1021/acssynbio.8b00052. Epub 2018 Aug 14. ACS Synth Biol. 2018. PMID: 30064218 Free PMC article.
-
Engineering allostery.Trends Genet. 2014 Dec;30(12):521-8. doi: 10.1016/j.tig.2014.09.004. Epub 2014 Oct 8. Trends Genet. 2014. PMID: 25306102 Free PMC article. Review.
-
Lactose repressor protein: functional properties and structure.Prog Nucleic Acid Res Mol Biol. 1998;58:127-64. doi: 10.1016/s0079-6603(08)60035-5. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9308365 Review.
Cited by
-
Step-by-step design of proteins for small molecule interaction: A review on recent milestones.Protein Sci. 2021 Aug;30(8):1502-1520. doi: 10.1002/pro.4098. Epub 2021 May 10. Protein Sci. 2021. PMID: 33934427 Free PMC article. Review.
-
Evolution-guided engineering of small-molecule biosensors.Nucleic Acids Res. 2020 Jan 10;48(1):e3. doi: 10.1093/nar/gkz954. Nucleic Acids Res. 2020. PMID: 31777933 Free PMC article.
-
Scaling up genetic circuit design for cellular computing: advances and prospects.Nat Comput. 2018;17(4):833-853. doi: 10.1007/s11047-018-9715-9. Epub 2018 Oct 5. Nat Comput. 2018. PMID: 30524216 Free PMC article.
-
Computational redesign of the Escherichia coli ribose-binding protein ligand binding pocket for 1,3-cyclohexanediol and cyclohexanol.Sci Rep. 2019 Nov 15;9(1):16940. doi: 10.1038/s41598-019-53507-5. Sci Rep. 2019. PMID: 31729460 Free PMC article.
-
Engineering Ag43 Signal Peptides with Bacterial Display and Selection.Methods Protoc. 2022 Dec 23;6(1):1. doi: 10.3390/mps6010001. Methods Protoc. 2022. PMID: 36648950 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

