Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus
- PMID: 26689811
- PMCID: PMC4687282
- DOI: 10.1186/s12985-015-0421-2
Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus
Erratum in
-
Erratum to: Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus.Virol J. 2016 Feb 1;13:19. doi: 10.1186/s12985-016-0465-y. Virol J. 2016. PMID: 26833094 Free PMC article. No abstract available.
Abstract
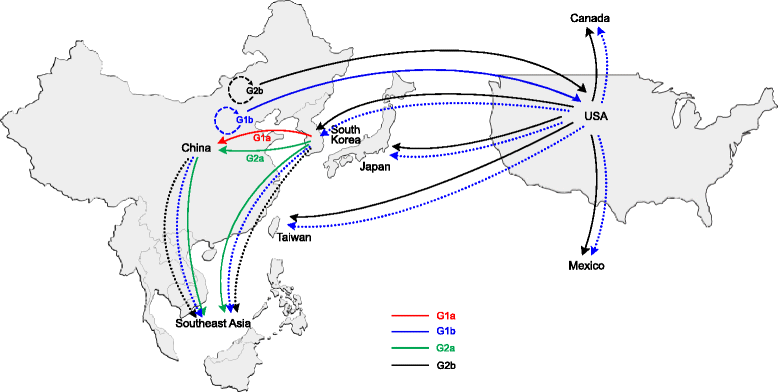
The enteric disease of swine recognized in the early 1970s in Europe was initially described as "epidemic viral diarrhea" and is now termed "porcine epidemic diarrhea (PED)". The coronavirus referred to as PED virus (PEDV) was determined to be the etiologic agent of this disease in the late 1970s. Since then the disease has been reported in Europe and Asia, but the most severe outbreaks have occurred predominantly in Asian swine-producing countries. Most recently, PED first emerged in early 2013 in the United States that caused high morbidity and mortality associated with PED, remarkably affecting US pig production, and spread further to Canada and Mexico. Soon thereafter, large-scale PED epidemics recurred through the pork industry in South Korea, Japan, and Taiwan. These recent outbreaks and global re-emergence of PED require urgent attention and deeper understanding of PEDV biology and pathogenic mechanisms. This paper highlights the current knowledge of molecular epidemiology, diagnosis, and pathogenesis of PEDV, as well as prevention and control measures against PEDV infection. More information about the virus and the disease is still necessary for the development of effective vaccines and control strategies. It is hoped that this review will stimulate further basic and applied studies and encourage collaboration among producers, researchers, and swine veterinarians to provide answers that improve our understanding of PEDV and PED in an effort to eliminate this economically significant viral disease, which emerged or re-emerged worldwide.
Figures
References
-
- Oldham J. Letter to the editor. Pig Farming. 1972;10:72–3.
-
- Debouck P, Pensaert M. Experimental infection of pigs with a new porcine enteric coronavirus, CV777. Am J Vet Res. 1980;41:219–23. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

