The impacts of polyploidy, geographic and ecological isolations on the diversification of Panax (Araliaceae)
- PMID: 26690782
- PMCID: PMC4687065
- DOI: 10.1186/s12870-015-0669-0
The impacts of polyploidy, geographic and ecological isolations on the diversification of Panax (Araliaceae)
Abstract
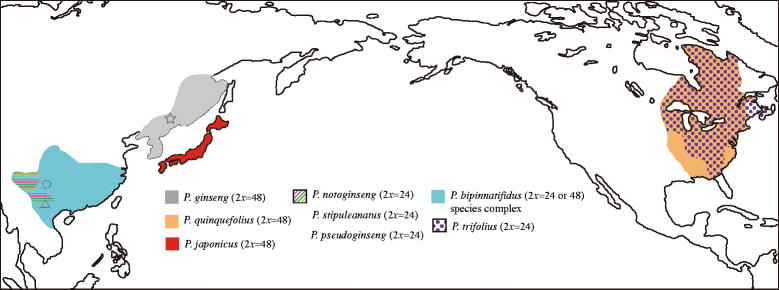
Background: Panax L. is a medicinally important genus within family Araliaceae, where almost all species are of cultural significance for traditional Chinese medicine. Previous studies suggested two independent origins of the East Asia and North America disjunct distribution of this genus and multiple rounds of whole genome duplications (WGDs) might have occurred during the evolutionary process.
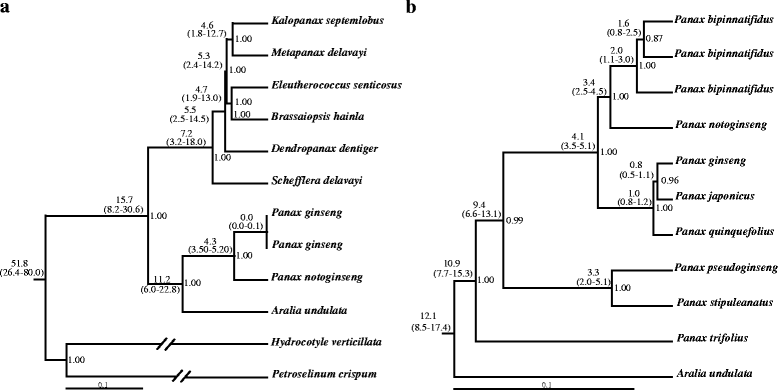
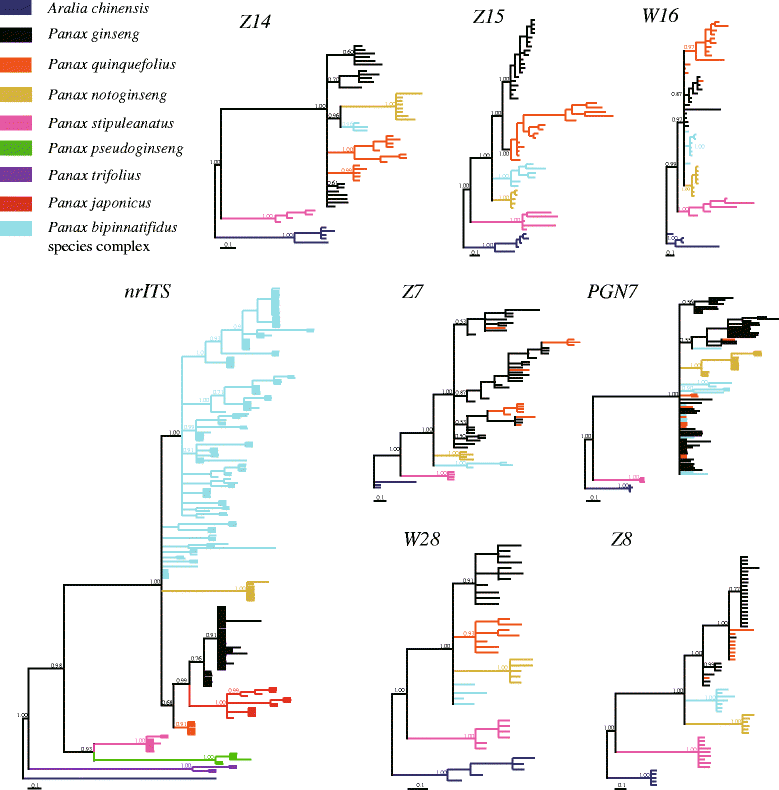
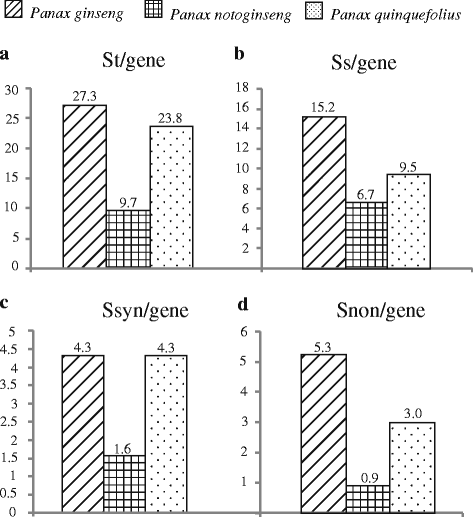
Results: We employed multiple chloroplast and nuclear markers to investigate the evolution and diversification of Panax. Our phylogenetic analyses confirmed previous observations of the independent origins of disjunct distribution and both ancient and recent WGDs have occurred within Panax. The estimations of divergence time implied that the ancient WGD might have occurred before the establishment of Panax. Thereafter, at least two independent recent WGD events have occurred within Panax, one of which has led to the formation of three geographically isolated tetraploid species P. ginseng, P. japonicus and P. quinquefolius. Population genetic analyses showed that the diploid species P. notoginseng harbored significantly lower nucleotide diversity than those of the two tetraploid species P. ginseng and P. quinquefolius and the three species showed distinct nucleotide variation patterns at exon regions.
Conclusion: Our findings based on the phylogenetic and population genetic analyses, coupled with the species distribution patterns of Panax, suggested that the two rounds of WGD along with the geographic and ecological isolations might have together contributed to the evolution and diversification of this genus.
Figures
References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

