Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway
- PMID: 26691337
- PMCID: PMC4687329
- DOI: 10.1186/s12934-015-0393-3
Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway
Abstract
Background: Heterologous expression systems based on promoters inducible with isopropyl-β-D-1-thiogalactopyranoside (IPTG), e.g., Escherichia coli BL21(DE3) and cognate LacI(Q)/P(lacUV5)-T7 vectors, are commonly used for production of recombinant proteins and metabolic pathways. The applicability of such cell factories is limited by the complex physiological burden imposed by overexpression of the exogenous genes during a bioprocess. This burden originates from a combination of stresses that may include competition for the expression machinery, side-reactions due to the activity of the recombinant proteins, or the toxicity of their substrates, products and intermediates. However, the physiological impact of IPTG-induced conditional expression on the recombinant host under such harsh conditions is often overlooked.
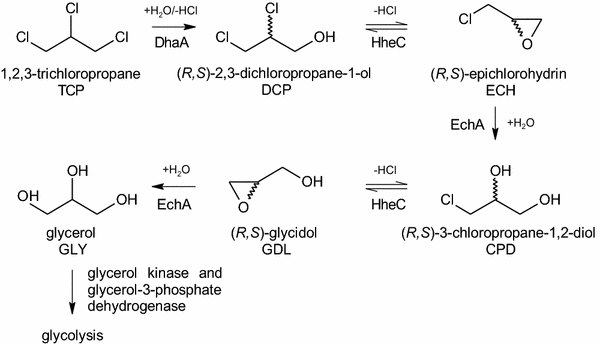
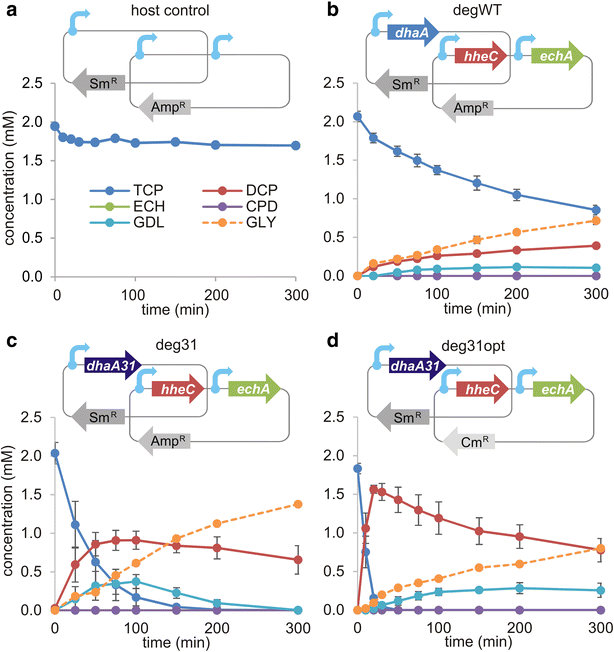
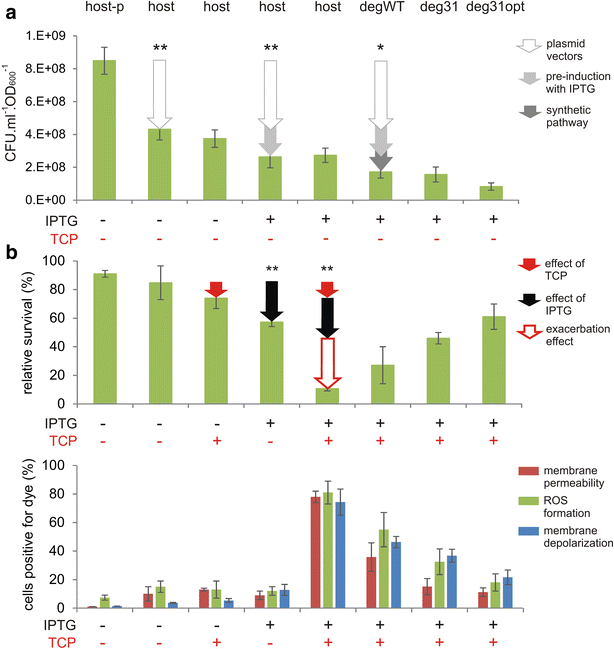
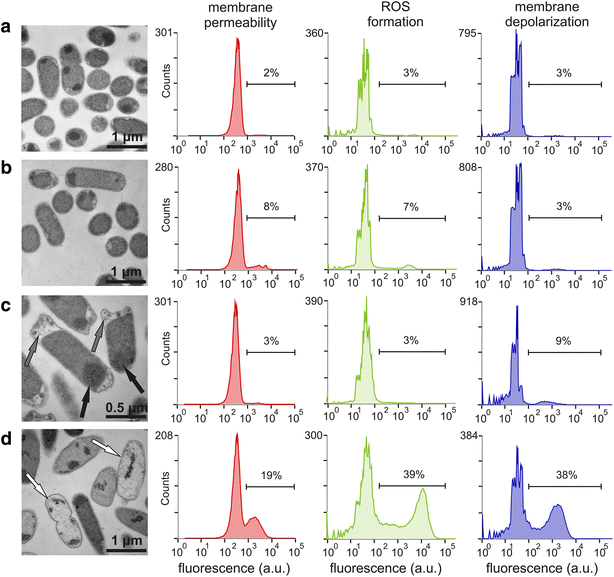
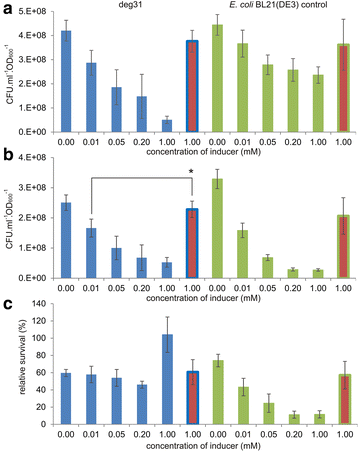
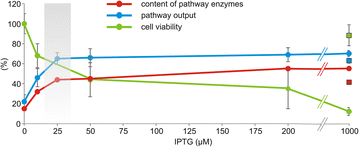
Results: The physiological responses to IPTG of the E. coli BL21(DE3) strain and three different recombinants carrying a synthetic metabolic pathway for biodegradation of the toxic anthropogenic pollutant 1,2,3-trichloropropane (TCP) were investigated using plating, flow cytometry, and electron microscopy. Collected data revealed unexpected negative synergistic effect of inducer of the expression system and toxic substrate resulting in pronounced physiological stress. Replacing IPTG with the natural sugar effector lactose greatly reduced such stress, demonstrating that the effect was due to the original inducer's chemical properties.
Conclusions: IPTG is not an innocuous inducer; instead, it exacerbates the toxicity of haloalkane substrate and causes appreciable damage to the E. coli BL21(DE3) host, which is already bearing a metabolic burden due to its content of plasmids carrying the genes of the synthetic metabolic pathway. The concentration of IPTG can be effectively tuned to mitigate this negative effect. Importantly, we show that induction with lactose, the natural inducer of P lac , dramatically lightens the burden without reducing the efficiency of the synthetic TCP degradation pathway. This suggests that lactose may be a better inducer than IPTG for the expression of heterologous pathways in E. coli BL21(DE3).
Figures
Similar articles
-
High-level production of membrane proteins in E. coli BL21(DE3) by omitting the inducer IPTG.Microb Cell Fact. 2015 Sep 16;14:142. doi: 10.1186/s12934-015-0328-z. Microb Cell Fact. 2015. PMID: 26377812 Free PMC article.
-
Development of inducer-free expression plasmids based on IPTG-inducible promoters for Bacillus subtilis.Microb Cell Fact. 2017 Jul 25;16(1):130. doi: 10.1186/s12934-017-0747-0. Microb Cell Fact. 2017. PMID: 28743271 Free PMC article.
-
E. coli HMS174(DE3) is a sustainable alternative to BL21(DE3).Microb Cell Fact. 2018 Oct 30;17(1):169. doi: 10.1186/s12934-018-1016-6. Microb Cell Fact. 2018. PMID: 30376846 Free PMC article.
-
Efficient heterologous expression of cellobiose 2-epimerase gene in Escherichia coli under the control of T7 lac promoter without addition of IPTG and lactose.Protein Expr Purif. 2024 Nov;223:106558. doi: 10.1016/j.pep.2024.106558. Epub 2024 Jul 27. Protein Expr Purif. 2024. PMID: 39074650
-
Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels.J Mol Biol. 1996 Jul 19;260(3):289-98. doi: 10.1006/jmbi.1996.0399. J Mol Biol. 1996. PMID: 8757792 Review.
Cited by
-
Boosting Recombinant Inclusion Body Production-From Classical Fed-Batch Approach to Continuous Cultivation.Front Bioeng Biotechnol. 2019 Oct 31;7:297. doi: 10.3389/fbioe.2019.00297. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31737617 Free PMC article.
-
Customized chitooligosaccharide production-controlling their length via engineering of rhizobial chitin synthases and the choice of expression system.Front Bioeng Biotechnol. 2022 Dec 14;10:1073447. doi: 10.3389/fbioe.2022.1073447. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36588959 Free PMC article.
-
Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non-traditional microorganisms.Microb Biotechnol. 2019 Jan;12(1):98-124. doi: 10.1111/1751-7915.13292. Epub 2018 Jun 21. Microb Biotechnol. 2019. PMID: 29926529 Free PMC article. Review.
-
Improving trans-cinnamic acid production in a model cyanobacterium.Biotechnol Prog. 2025 Mar-Apr;41(2):e3512. doi: 10.1002/btpr.3512. Epub 2024 Oct 1. Biotechnol Prog. 2025. PMID: 40235106 Free PMC article.
-
Heterologous expression and structure prediction of a xylanase identified from a compost metagenomic library.Appl Microbiol Biotechnol. 2024 May 10;108(1):329. doi: 10.1007/s00253-024-13169-4. Appl Microbiol Biotechnol. 2024. PMID: 38727750 Free PMC article.
References
-
- Choi JH, Keum KC, Lee SY. Production of recombinant proteins by high cell density culture of Escherichia coli. Chem Eng Sci. 2006;61(3):876–885. doi: 10.1016/j.ces.2005.03.031. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

