Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity
- PMID: 26691752
- PMCID: PMC4816136
- DOI: 10.1038/cr.2015.149
Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity
Erratum in
-
Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity.Cell Res. 2016 Aug;26(8):967. doi: 10.1038/cr.2016.90. Cell Res. 2016. PMID: 27481604 Free PMC article. No abstract available.
Abstract
Sensory neurons are distinguished by distinct signaling networks and receptive characteristics. Thus, sensory neuron types can be defined by linking transcriptome-based neuron typing with the sensory phenotypes. Here we classify somatosensory neurons of the mouse dorsal root ganglion (DRG) by high-coverage single-cell RNA-sequencing (10 950 ± 1 218 genes per neuron) and neuron size-based hierarchical clustering. Moreover, single DRG neurons responding to cutaneous stimuli are recorded using an in vivo whole-cell patch clamp technique and classified by neuron-type genetic markers. Small diameter DRG neurons are classified into one type of low-threshold mechanoreceptor and five types of mechanoheat nociceptors (MHNs). Each of the MHN types is further categorized into two subtypes. Large DRG neurons are categorized into four types, including neurexophilin 1-expressing MHNs and mechanical nociceptors (MNs) expressing BAI1-associated protein 2-like 1 (Baiap2l1). Mechanoreceptors expressing trafficking protein particle complex 3-like and Baiap2l1-marked MNs are subdivided into two subtypes each. These results provide a new system for cataloging somatosensory neurons and their transcriptome databases.
Figures

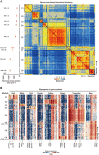
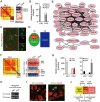



References
-
- Maxwell DJ, Réthelyi M. Ultrastructure and synaptic connections of cutaneius afferent fibres in the spinal cord. Trends Neurosci 1987; 10:117–123.
-
- Ju G, Hökfelt T, Brodin E, et al. Primary sensory neurons of the rat showing calcitonin gene-related peptide immunoreactivity and their relation to substance P-, somatostatin-, galanin-, vasoactive intestinal polypeptide- and cholecystokinin-immunoreactive ganglion cells. Cell Tissue Res 1987; 247:417–431. - PubMed
-
- Wang H, Rivero-Melian C, Robertson B, Grant G. Transganglionic transport and binding of the isolectin B4 from Griffonia simplicifolia I in rat primary sensory neurons. Neuroscience 1994; 62:539–551. - PubMed
-
- Meyer RA, Ringkamp M, Campbell JN, Raja SN. Peripheral mechanisms of cutaneous nociception. In: McMahon SB, Koltzenburg M. eds. Wall and Melzack's Textbook of Pain. Elsevier 2006:3–34.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

