Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein
- PMID: 26691874
- PMCID: PMC7108309
- DOI: 10.1016/j.drudis.2015.11.015
Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein
Abstract
The advent of severe acute respiratory syndrome (SARS) in the 21st century and the recent outbreak of Middle-East respiratory syndrome (MERS) highlight the importance of coronaviruses (CoVs) as human pathogens, emphasizing the need for development of novel antiviral strategies to combat acute respiratory infections caused by CoVs. Recent studies suggest that nucleocapsid (N) proteins from coronaviruses and other viruses can be useful antiviral drug targets against viral infections. This review aims to provide readers with a concise survey of the structural features of coronavirus N proteins and how these features provide insights into structure-based development of therapeutics against coronaviruses. We will also present our latest results on MERS-CoV N protein and its potential as an antiviral drug target.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

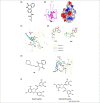
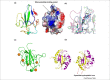
Similar articles
-
MOV10 Helicase Interacts with Coronavirus Nucleocapsid Protein and Has Antiviral Activity.mBio. 2021 Oct 26;12(5):e0131621. doi: 10.1128/mBio.01316-21. Epub 2021 Sep 14. mBio. 2021. PMID: 34517762 Free PMC article.
-
Recent progress in the discovery of inhibitors targeting coronavirus proteases.Virol Sin. 2016 Feb;31(1):24-30. doi: 10.1007/s12250-015-3711-3. Epub 2016 Feb 19. Virol Sin. 2016. PMID: 26920707 Free PMC article. Review.
-
Severe acute respiratory syndrome vs. the Middle East respiratory syndrome.Curr Opin Pulm Med. 2014 May;20(3):233-41. doi: 10.1097/MCP.0000000000000046. Curr Opin Pulm Med. 2014. PMID: 24626235 Review.
-
Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus.J Infect Public Health. 2016 May-Jun;9(3):227-30. doi: 10.1016/j.jiph.2016.04.001. Epub 2016 Apr 16. J Infect Public Health. 2016. PMID: 27095301 Free PMC article. Review.
-
Advances in MERS-CoV Vaccines and Therapeutics Based on the Receptor-Binding Domain.Viruses. 2019 Jan 14;11(1):60. doi: 10.3390/v11010060. Viruses. 2019. PMID: 30646569 Free PMC article. Review.
Cited by
-
Antiviral activity of green tea and black tea polyphenols in prophylaxis and treatment of COVID-19: A review.Phytomedicine. 2021 May;85:153286. doi: 10.1016/j.phymed.2020.153286. Epub 2020 Jul 17. Phytomedicine. 2021. PMID: 32741697 Free PMC article. Review.
-
Targeting Intrinsically Disordered Proteins through Dynamic Interactions.Biomolecules. 2020 May 11;10(5):743. doi: 10.3390/biom10050743. Biomolecules. 2020. PMID: 32403216 Free PMC article. Review.
-
In silico investigation of phytoconstituents from Indian medicinal herb 'Tinospora cordifolia (giloy)' against SARS-CoV-2 (COVID-19) by molecular dynamics approach.J Biomol Struct Dyn. 2021 Oct;39(17):6792-6809. doi: 10.1080/07391102.2020.1803968. Epub 2020 Aug 7. J Biomol Struct Dyn. 2021. PMID: 32762511 Free PMC article.
-
Exploring the Targets of Novel Corona Virus and Docking-based Screening of Potential Natural Inhibitors to Combat COVID-19.Curr Top Med Chem. 2022;22(29):2410-2434. doi: 10.2174/1568026623666221020163831. Curr Top Med Chem. 2022. PMID: 36281864
-
Surface Plasmon Resonance Immunosensor for Direct Detection of Antibodies against SARS-CoV-2 Nucleocapsid Protein.Int J Mol Sci. 2024 Aug 6;25(16):8574. doi: 10.3390/ijms25168574. Int J Mol Sci. 2024. PMID: 39201259 Free PMC article.
References
-
- Cherry D.K. National Ambulatory Medical Care Survey: 2006 summary. Natl. Health Stat. Report. 2008;3:1–39. - PubMed
-
- Hsiao C-J. National Ambulatory Medical Care Survey: 2007 summary. Natl. Health Stat. Report. 2010;27:1–32. - PubMed
-
- Matoba Y. Detection of the human coronavirus 229E, HKU1, NL63, and OC43 between 2010 and 2013 in Yamagata, Japan. Jpn. J. Infect. Dis. 2015;68:138–141. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

