A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants
- PMID: 26691988
- PMCID: PMC4745342
- DOI: 10.1038/ng.3448
A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants
Abstract
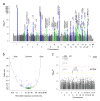
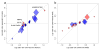
Advanced age-related macular degeneration (AMD) is the leading cause of blindness in the elderly, with limited therapeutic options. Here we report on a study of >12 million variants, including 163,714 directly genotyped, mostly rare, protein-altering variants. Analyzing 16,144 patients and 17,832 controls, we identify 52 independently associated common and rare variants (P < 5 × 10(-8)) distributed across 34 loci. Although wet and dry AMD subtypes exhibit predominantly shared genetics, we identify the first genetic association signal specific to wet AMD, near MMP9 (difference P value = 4.1 × 10(-10)). Very rare coding variants (frequency <0.1%) in CFH, CFI and TIMP3 suggest causal roles for these genes, as does a splice variant in SLC16A8. Our results support the hypothesis that rare coding variants can pinpoint causal genes within known genetic loci and illustrate that applying the approach systematically to detect new loci requires extremely large sample sizes.
Figures


References
-
- Smith W, et al. Risk factors for age-related macular degeneration: Pooled findings from three continents. Ophthalmology. 2001;108:697–704. - PubMed
-
- Chakravarthy U, Evans J, Rosenfeld PJ. Age related macular degeneration. BMJ. 2010;340:c981. - PubMed
-
- Wong WL, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2:e106–16. - PubMed
Publication types
MeSH terms
Grants and funding
- K22 LM011938/LM/NLM NIH HHS/United States
- R01 EY013435/EY/NEI NIH HHS/United States
- P30 EY014801/EY/NEI NIH HHS/United States
- UL1 TR000427/TR/NCATS NIH HHS/United States
- T32 GM080178/GM/NIGMS NIH HHS/United States
- P30 DK079626/DK/NIDDK NIH HHS/United States
- R01 EY022310/EY/NEI NIH HHS/United States
- P30 EY019007/EY/NEI NIH HHS/United States
- F31 AG044089/AG/NIA NIH HHS/United States
- 1X01HG006934-01/HG/NHGRI NIH HHS/United States
- HHSN268201200008I/HL/NHLBI NIH HHS/United States
- T32 EY023194/EY/NEI NIH HHS/United States
- U54 HG006542/HG/NHGRI NIH HHS/United States
- T32 EY007157/EY/NEI NIH HHS/United States
- U10 EY006594/EY/NEI NIH HHS/United States
- R01 AG019085/AG/NIA NIH HHS/United States
- R01 EY012118/EY/NEI NIH HHS/United States
- HHSN268201200008C/HL/NHLBI NIH HHS/United States
- U01 HG006389/HG/NHGRI NIH HHS/United States
- R01 EY021532/EY/NEI NIH HHS/United States
- R01 EY023164/EY/NEI NIH HHS/United States
- MC_PC_U127561128/MRC_/Medical Research Council/United Kingdom
- P30 EY001583/EY/NEI NIH HHS/United States
- IK6 BX005233/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

