Upstream AUGs and upstream ORFs can regulate the downstream ORF in Plasmodium falciparum
- PMID: 26692187
- PMCID: PMC4687322
- DOI: 10.1186/s12936-015-1040-5
Upstream AUGs and upstream ORFs can regulate the downstream ORF in Plasmodium falciparum
Abstract
Background: Upstream open reading frames (uORFs) and upstream AUGs (uAUGs) can regulate the translation of downstream ORFs. The AT rich genome of Plasmodium falciparum, due to the higher AT content of start and stop codons, has the potential to give rise to a large number of uORFs and uAUGs that may affect expression of their flanking ORFs.
Methods: A bioinformatics approach was used to detect uATGs associated with different genes in the parasite. To study the effect of some of these uAUGs on the expression of the downstream ORF, promoters and 5' leaders containing uAUGs and uORFs were cloned upstream of a luciferase reporter gene. Luciferase assays were carried out in transient transfection experiments to assess the effects of uAUGs and mutations on reporter expression.
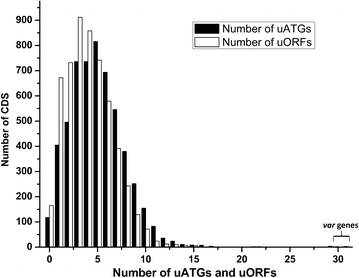
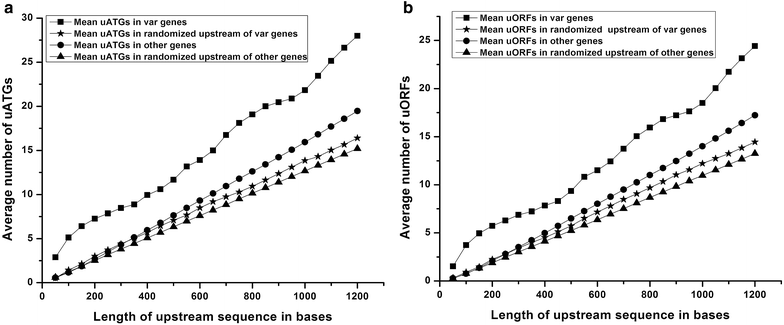
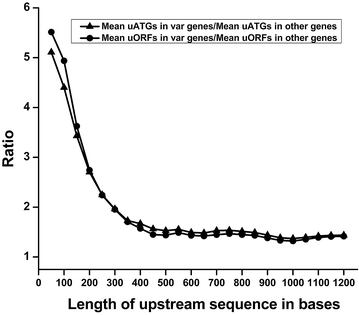
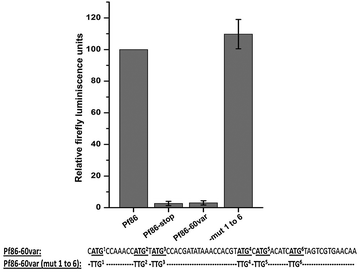
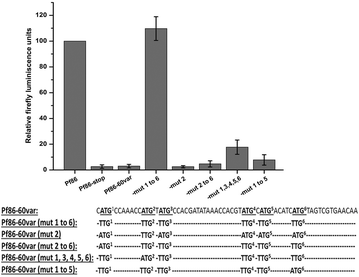
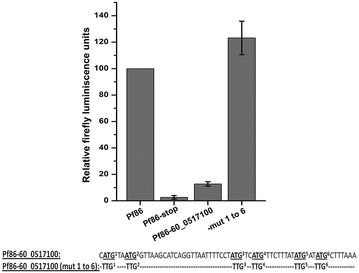
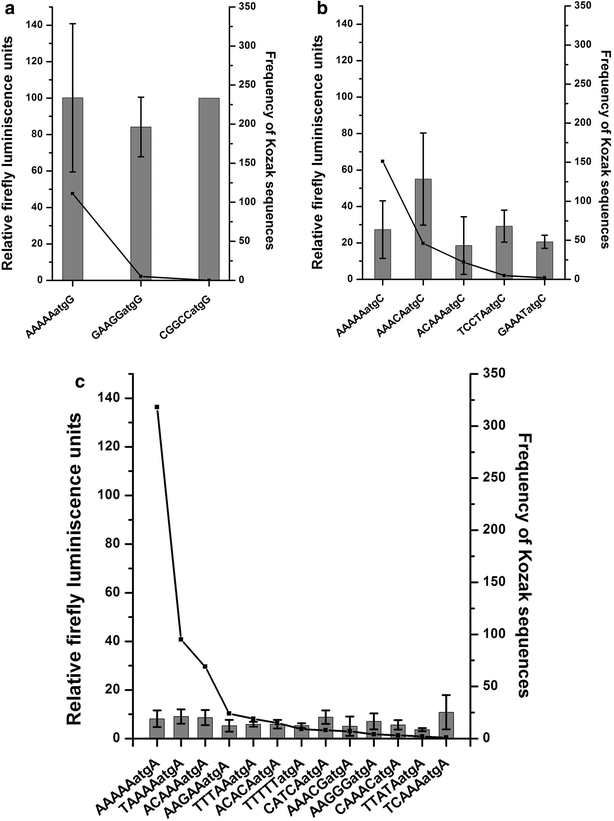
Results: The average number of uATGs and uORFs seen in P. falciparum coding sequences (CDS) is expectedly high compared to other less biased genomes. Certain genes, including the var gene family contain the maximum number of uATGs and uORFs in the parasite. They possess ~5 times more uORFs and ~4.5 times more uAUGs within 100 bases upstream of the start codons than other CDS of the parasite. A 60 bp upstream region containing three ORFs and five ATGs from var gene PF3D7_0400100 and a gene of unknown function (PF3D7_0517100) when cloned upstream of the luciferase start codon, driven by the hsp86 promoter, resulted in loss of luciferase activity. This was restored when all the ATGs present in the -60 bp were mutated to TTGs. Point mutations in the ATGs showed that even one AUG was sufficient to repress the luciferase gene.
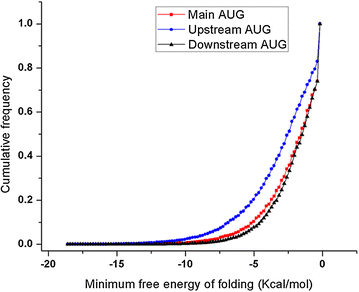
Conclusions: Overall, this work indicates that the P. falciparum genome has a large number of uATGs and uORFs that can repress the expression of flanking ORFs. The role of AUGs in translation initiation suggests that this repression is mediated by preventing the translation initiation complex from reaching the main AUG of the downstream ORF. How the P. falciparum ribosome is able to bypass these uAUGs and uORFs for highly expressed genes remains a question for future research.
Figures
Similar articles
-
5'-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation.Nucleic Acids Res. 2004 Feb 27;32(4):1382-91. doi: 10.1093/nar/gkh305. Print 2004. Nucleic Acids Res. 2004. PMID: 14990743 Free PMC article.
-
Messenger RNAs with large numbers of upstream open reading frames are translated via leaky scanning and reinitiation in the asexual stages of Plasmodium falciparum.Parasitology. 2020 Sep;147(10):1100-1113. doi: 10.1017/S0031182020000840. Epub 2020 May 26. Parasitology. 2020. PMID: 32450933 Free PMC article.
-
Rare codons in uORFs of baculovirus p13 gene modulates downstream gene expression.Virus Res. 2011 Jan;155(1):249-53. doi: 10.1016/j.virusres.2010.10.016. Epub 2010 Oct 21. Virus Res. 2011. PMID: 20970467
-
The role of upstream open reading frames in translation regulation in the apicomplexan parasites Plasmodium falciparum and Toxoplasma gondii.Parasitology. 2021 Sep;148(11):1277-1287. doi: 10.1017/S0031182021000937. Epub 2021 Jun 8. Parasitology. 2021. PMID: 34099078 Free PMC article. Review.
-
Translational Regulation by Upstream Open Reading Frames and Human Diseases.Adv Exp Med Biol. 2019;1157:99-116. doi: 10.1007/978-3-030-19966-1_5. Adv Exp Med Biol. 2019. PMID: 31342439 Review.
Cited by
-
Functional characterization of 5' UTR cis-acting sequence elements that modulate translational efficiency in Plasmodium falciparum and humans.Malar J. 2022 Jan 6;21(1):15. doi: 10.1186/s12936-021-04024-2. Malar J. 2022. PMID: 34991611 Free PMC article.
-
Recent advances in malaria genomics and epigenomics.Genome Med. 2016 Sep 7;8(1):92. doi: 10.1186/s13073-016-0343-7. Genome Med. 2016. PMID: 27605022 Free PMC article. Review.
-
Deciphering Transcription in Cryptosporidium parvum: Polycistronic Gene Expression and Chromatin Accessibility.bioRxiv [Preprint]. 2025 Jan 18:2025.01.17.633476. doi: 10.1101/2025.01.17.633476. bioRxiv. 2025. PMID: 39868316 Free PMC article. Preprint.
-
Translational regulation in blood stages of the malaria parasite Plasmodium spp.: systems-wide studies pave the way.Wiley Interdiscip Rev RNA. 2016 Nov;7(6):772-792. doi: 10.1002/wrna.1365. Epub 2016 May 26. Wiley Interdiscip Rev RNA. 2016. PMID: 27230797 Free PMC article. Review.
-
Regulation of PfEMP1-VAR2CSA translation by a Plasmodium translation-enhancing factor.Nat Microbiol. 2017 May 8;2:17068. doi: 10.1038/nmicrobiol.2017.68. Nat Microbiol. 2017. PMID: 28481333
References
-
- WHO. World Malaria Report 2014. Geneva: World Health Organization; 2014.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

