Interleukin-1 antagonism moderates the inflammatory state associated with Type 1 diabetes during clinical trials conducted at disease onset
- PMID: 26692253
- PMCID: PMC4828314
- DOI: 10.1002/eji.201546005
Interleukin-1 antagonism moderates the inflammatory state associated with Type 1 diabetes during clinical trials conducted at disease onset
Abstract
It was hypothesized that IL-1 antagonism would preserve β-cell function in new onset Type 1 diabetes (T1D). However, the Anti-Interleukin-1 in Diabetes Action (AIDA) and TrialNet Canakinumab (TN-14) trials failed to show efficacy of IL-1 receptor antagonist (IL-1Ra) or canakinumab, as measured by stimulated C-peptide response. Additional measures are needed to define immune state changes associated with therapeutic responses. Here, we studied these trial participants with plasma-induced transcriptional analysis. In blinded analyses, 70.2% of AIDA and 68.9% of TN-14 participants were correctly called to their treatment arm. While the transcriptional signatures from the two trials were distinct, both therapies achieved varying immunomodulation consistent with IL-1 inhibition. On average, IL-1 antagonism resulted in modest normalization relative to healthy controls. At endpoint, signatures were quantified using a gene ontology-based inflammatory index, and an inverse relationship was observed between measured inflammation and stimulated C-peptide response in IL-1Ra- and canakinumab-treated patients. Cytokine neutralization studies showed that IL-1α and IL-1β additively contribute to the T1D inflammatory state. Finally, analyses of baseline signatures were indicative of later therapeutic response. Despite the absence of clinical efficacy by IL-1 antagonist therapy, transcriptional analysis detected immunomodulation and may yield new insight when applied to other clinical trials.
Keywords: Anakinra; Canakinumab; Interleukin-1; Transcriptional signatures; Type 1 diabetes.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Figures
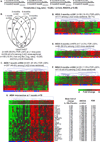

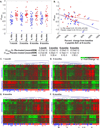


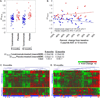


References
-
- Mandrup-Poulsen T, Pickersgill L, Donath MY. Blockade of interleukin 1 in type 1 diabetes mellitus. Nat Rev Endocrinol. 2010;6:158–166. - PubMed
-
- Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. - PubMed
-
- Stiller CR, Dupre J, Gent M, Jenner MR, Keown PA, Laupacis A, Martell R, Rodger NW, von Graffenried B, Wolfe BM. Effects of cyclosporine immunosuppression in insulin-dependent diabetes mellitus of recent onset. Science. 1984;223:1362–1367. - PubMed
-
- Keymeulen B, Vandemeulebroucke E, Ziegler AG, Mathieu C, Kaufman L, Hale G, Gorus F, Goldman M, Walter M, Candon S, Schandene L, Crenier L, De Block C, Seigneurin JM, De Pauw P, Pierard D, Weets I, Rebello P, Bird P, Berrie E, Frewin M, Waldmann H, Bach JF, Pipeleers D, Chatenoud L. Insulin needs after CD3-antibody therapy in new-onset type 1 diabetes. N Engl J Med. 2005;352:2598–2608. - PubMed
-
- Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, Becker DJ, Gitelman SE, Goland R, Gottlieb PA, Marks JB, McGee PF, Moran AM, Raskin P, Rodriguez H, Schatz DA, Wherrett D, Wilson DM, Lachin JM, Skyler JS. Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N Engl J Med. 2009;361:2143–2152. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 RR024139/RR/NCRR NIH HHS/United States
- U01DK085461/DK/NIDDK NIH HHS/United States
- R01DK080100/DK/NIDDK NIH HHS/United States
- U01 DK061055/DK/NIDDK NIH HHS/United States
- UL1 TR001427/TR/NCATS NIH HHS/United States
- U01DK085505/DK/NIDDK NIH HHS/United States
- U01 DK061036/DK/NIDDK NIH HHS/United States
- U01 DK061041/DK/NIDDK NIH HHS/United States
- U01 DK085476/DK/NIDDK NIH HHS/United States
- R01 DK080100/DK/NIDDK NIH HHS/United States
- M01 RR000400/RR/NCRR NIH HHS/United States
- 8UL1TR000055/TR/NCATS NIH HHS/United States
- U01 DK061010/DK/NIDDK NIH HHS/United States
- U01 DK084565/DK/NIDDK NIH HHS/United States
- UL1 RR025761/RR/NCRR NIH HHS/United States
- U01 DK061040/DK/NIDDK NIH HHS/United States
- UL1 RR024153/RR/NCRR NIH HHS/United States
- UL1 RR029890/RR/NCRR NIH HHS/United States
- UL1 TR001105/TR/NCATS NIH HHS/United States
- UL1 RR031986/RR/NCRR NIH HHS/United States
- U01 DK085466/DK/NIDDK NIH HHS/United States
- U01 DK103153/DK/NIDDK NIH HHS/United States
- U01 DK061058/DK/NIDDK NIH HHS/United States
- U01 DK085505/DK/NIDDK NIH HHS/United States
- U01 DK085453/DK/NIDDK NIH HHS/United States
- U01DK061055/DK/NIDDK NIH HHS/United States
- UL1 RR025780/RR/NCRR NIH HHS/United States
- M01 RR00400/RR/NCRR NIH HHS/United States
- U01 DK085499/DK/NIDDK NIH HHS/United States
- UL1 RR024131/RR/NCRR NIH HHS/United States
- U01 DK106984/DK/NIDDK NIH HHS/United States
- U01 DK085463/DK/NIDDK NIH HHS/United States
- UL1 TR001108/TR/NCATS NIH HHS/United States
- U01DK061036/DK/NIDDK NIH HHS/United States
- R01 AI078713/AI/NIAID NIH HHS/United States
- UL1 RR024982/RR/NCRR NIH HHS/United States
- R01AI078713/AI/NIAID NIH HHS/United States
- DP3DK098161/DK/NIDDK NIH HHS/United States
- U01 DK061016/DK/NIDDK NIH HHS/United States
- UL1 RR025744/RR/NCRR NIH HHS/United States
- U01 DK107014/DK/NIDDK NIH HHS/United States
- DP3 DK098161/DK/NIDDK NIH HHS/United States
- U01 DK061042/DK/NIDDK NIH HHS/United States
- UL1 TR000055/TR/NCATS NIH HHS/United States
- U01 DK061034/DK/NIDDK NIH HHS/United States
- U01 DK085461/DK/NIDDK NIH HHS/United States
- U01 DK085509/DK/NIDDK NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- HHSN267200800019C/LM/NLM NIH HHS/United States
- U01 DK085465/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

