Genetic Architecture of Atherosclerosis in Mice: A Systems Genetics Analysis of Common Inbred Strains
- PMID: 26694027
- PMCID: PMC4687930
- DOI: 10.1371/journal.pgen.1005711
Genetic Architecture of Atherosclerosis in Mice: A Systems Genetics Analysis of Common Inbred Strains
Erratum in
-
Correction: Genetic Architecture of Atherosclerosis in Mice: A Systems Genetics Analysis of Common Inbred Strains.PLoS Genet. 2016 Mar 2;12(3):e1005913. doi: 10.1371/journal.pgen.1005913. eCollection 2016 Mar. PLoS Genet. 2016. PMID: 26934746 Free PMC article. No abstract available.
Abstract
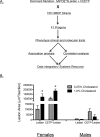
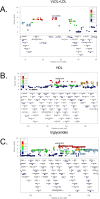
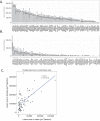
Common forms of atherosclerosis involve multiple genetic and environmental factors. While human genome-wide association studies have identified numerous loci contributing to coronary artery disease and its risk factors, these studies are unable to control environmental factors or examine detailed molecular traits in relevant tissues. We now report a study of natural variations contributing to atherosclerosis and related traits in over 100 inbred strains of mice from the Hybrid Mouse Diversity Panel (HMDP). The mice were made hyperlipidemic by transgenic expression of human apolipoprotein E-Leiden (APOE-Leiden) and human cholesteryl ester transfer protein (CETP). The mice were examined for lesion size and morphology as well as plasma lipid, insulin and glucose levels, and blood cell profiles. A subset of mice was studied for plasma levels of metabolites and cytokines. We also measured global transcript levels in aorta and liver. Finally, the uptake of acetylated LDL by macrophages from HMDP mice was quantitatively examined. Loci contributing to the traits were mapped using association analysis, and relationships among traits were examined using correlation and statistical modeling. A number of conclusions emerged. First, relationships among atherosclerosis and the risk factors in mice resemble those found in humans. Second, a number of trait-loci were identified, including some overlapping with previous human and mouse studies. Third, gene expression data enabled enrichment analysis of pathways contributing to atherosclerosis and prioritization of candidate genes at associated loci in both mice and humans. Fourth, the data provided a number of mechanistic inferences; for example, we detected no association between macrophage uptake of acetylated LDL and atherosclerosis. Fifth, broad sense heritability for atherosclerosis was much larger than narrow sense heritability, indicating an important role for gene-by-gene interactions. Sixth, stepwise linear regression showed that the combined variations in plasma metabolites, including LDL/VLDL-cholesterol, trimethylamine N-oxide (TMAO), arginine, glucose and insulin, account for approximately 30 to 40% of the variation in atherosclerotic lesion area. Overall, our data provide a rich resource for studies of complex interactions underlying atherosclerosis.
Conflict of interest statement
Authors MY, TK and PSG are employees and shareholders of Bristol-Myers Squibb.
Figures
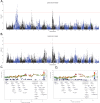



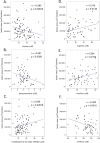

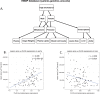
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- K99HL121172/HL/NHLBI NIH HHS/United States
- P01 HL28481/HL/NHLBI NIH HHS/United States
- K99/R00 HL102223/HL/NHLBI NIH HHS/United States
- HL103866/HL/NHLBI NIH HHS/United States
- P01 HL028481/HL/NHLBI NIH HHS/United States
- P20 HL113452/HL/NHLBI NIH HHS/United States
- T32 HL007895/HL/NHLBI NIH HHS/United States
- HL30568 NIH/NHLBI/PHS HHS/United States
- K99 HL121172/HL/NHLBI NIH HHS/United States
- R00 HL102223/HL/NHLBI NIH HHS/United States
- R00 HL121172/HL/NHLBI NIH HHS/United States
- P01 HL030568/HL/NHLBI NIH HHS/United States
- K99 HL102223/HL/NHLBI NIH HHS/United States
- R01 HL103866/HL/NHLBI NIH HHS/United States
- R01 HL128572/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

