Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum
- PMID: 26694030
- PMCID: PMC4703832
- DOI: 10.1038/ncomms10111
Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum
Abstract
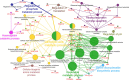
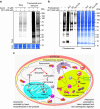
The mechanism of action of artemisinin and its derivatives, the most potent of the anti-malarial drugs, is not completely understood. Here we present an unbiased chemical proteomics analysis to directly explore this mechanism in Plasmodium falciparum. We use an alkyne-tagged artemisinin analogue coupled with biotin to identify 124 artemisinin covalent binding protein targets, many of which are involved in the essential biological processes of the parasite. Such a broad targeting spectrum disrupts the biochemical landscape of the parasite and causes its death. Furthermore, using alkyne-tagged artemisinin coupled with a fluorescent dye to monitor protein binding, we show that haem, rather than free ferrous iron, is predominantly responsible for artemisinin activation. The haem derives primarily from the parasite's haem biosynthesis pathway at the early ring stage and from haemoglobin digestion at the latter stages. Our results support a unifying model to explain the action and specificity of artemisinin in parasite killing.
Figures



Comment in
-
Chemical proteomics approach reveals the direct targets and the heme-dependent activation mechanism of artemisinin in Plasmodium falciparum using an artemisinin-based activity probe.Microb Cell. 2016 Apr 5;3(5):230-231. doi: 10.15698/mic2016.05.503. Microb Cell. 2016. PMID: 28357359 Free PMC article.
References
-
- Nosten F. & White N. J. Artemisinin-based combination treatment of falciparum malaria. Am. J. Trop. Med. Hyg. 77, 181–192 (2007). - PubMed
-
- Meshnick S. R. Artemisinin: mechanisms of action, resistance and toxicity. Int. J. Parasitol. 32, 1655–1660 (2002). - PubMed
-
- Meunier B. & Robert A. Heme as trigger and target for trioxane-containing antimalarial drugs. Acc. Chem. Res. 43, 1444–1451 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

