Meta-analysis of RNA-seq expression data across species, tissues and studies
- PMID: 26694591
- PMCID: PMC4699362
- DOI: 10.1186/s13059-015-0853-4
Meta-analysis of RNA-seq expression data across species, tissues and studies
Abstract
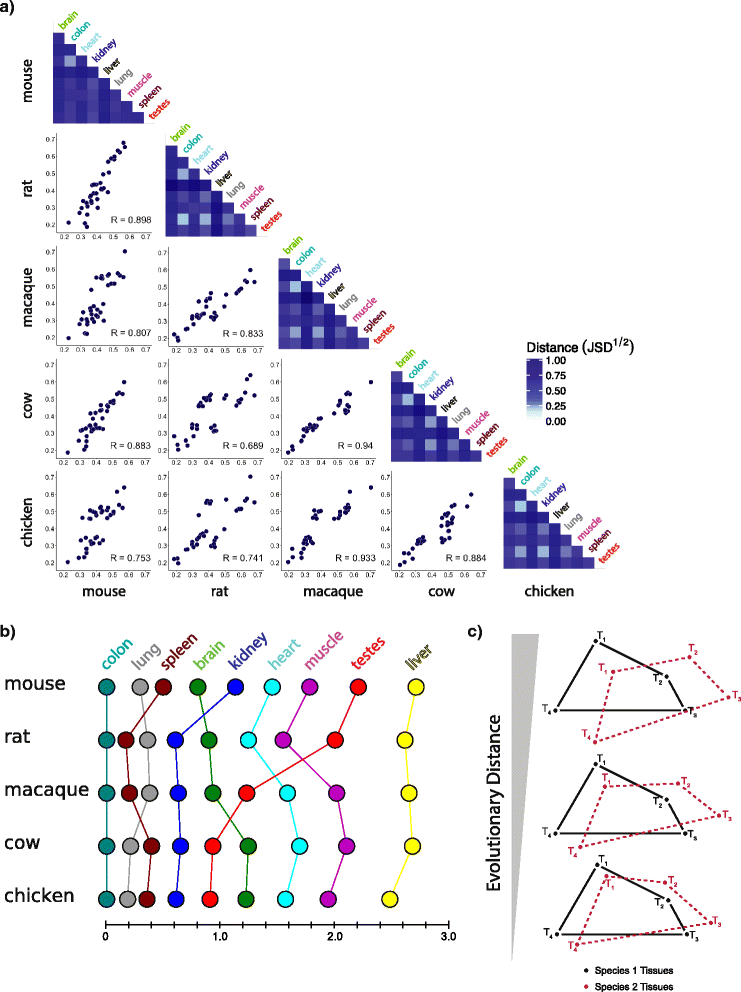
Background: Differences in gene expression drive phenotypic differences between species, yet major organs and tissues generally have conserved gene expression programs. Several comparative transcriptomic studies have observed greater similarity in gene expression between homologous tissues from different vertebrate species than between diverse tissues of the same species. However, a recent study by Lin and colleagues reached the opposite conclusion. These studies differed in the species and tissues analyzed, and in technical details of library preparation, sequencing, read mapping, normalization, gene sets, and clustering methods.
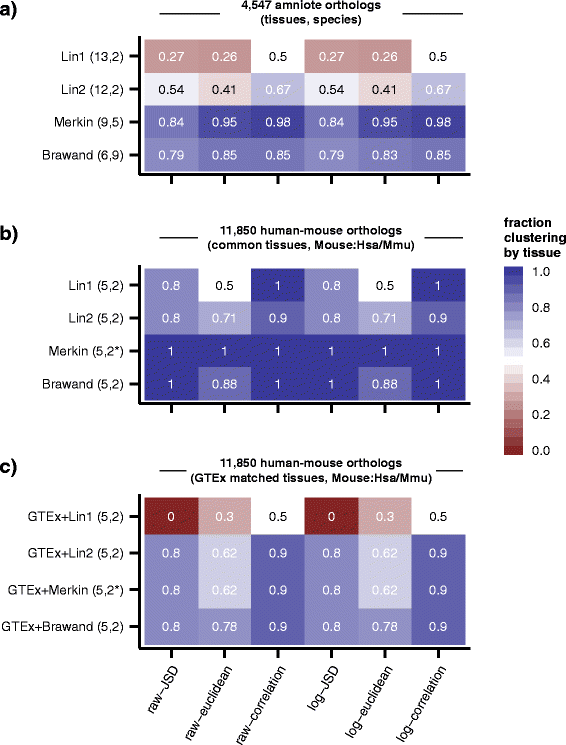
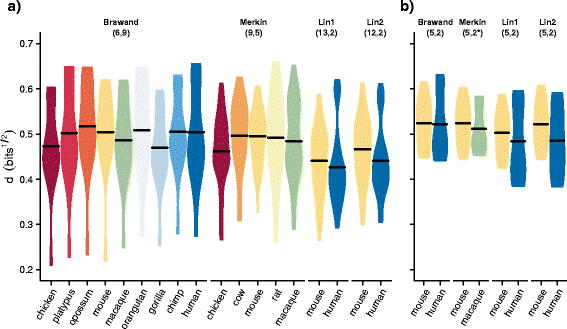
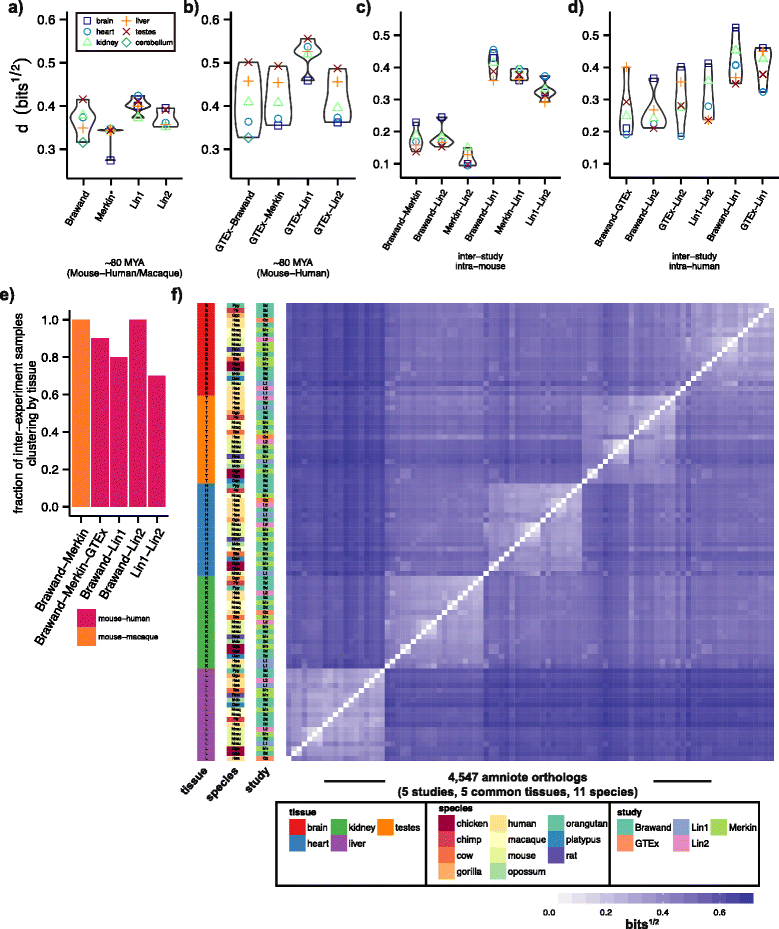
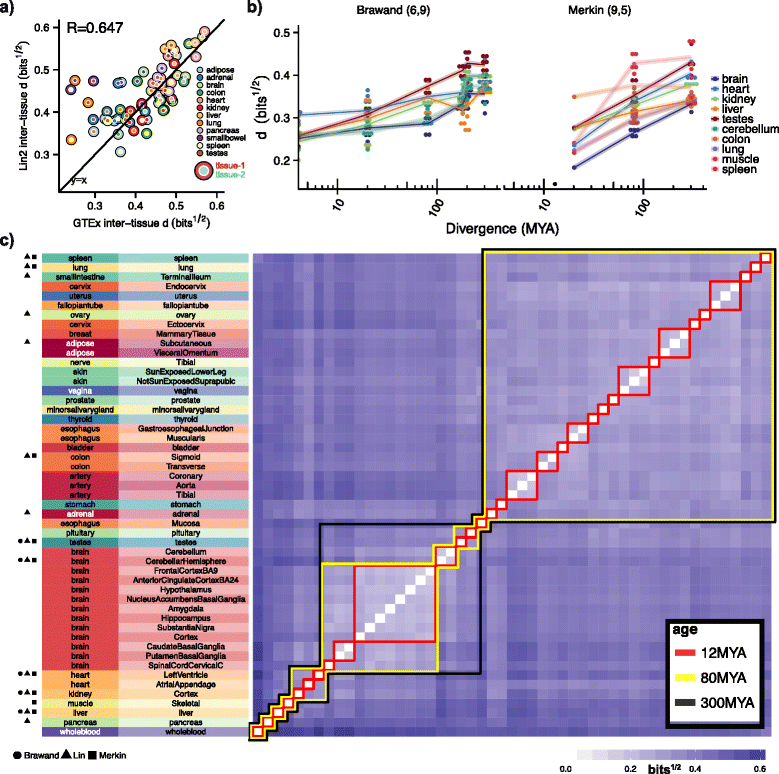
Results: To better understand gene expression evolution we reanalyzed data from four studies, including that of Lin, encompassing 6-13 tissues each from 11 vertebrate species using standardized mapping, normalization, and clustering methods. An analysis of independent data showed that the set of tissues chosen by Lin et al. were more similar to each other than those analyzed by previous studies. Comparing expression in five common tissues from the four studies, we observed that samples clustered exclusively by tissue rather than by species or study, supporting conservation of organ physiology in mammals. Furthermore, inter-study distances between homologous tissues were generally less than intra-study distances among different tissues, enabling informative meta-analyses. Notably, when comparing expression divergence of tissues over time to expression variation across 51 human GTEx tissues, we could accurately predict the clustering of expression for arbitrary pairs of tissues and species.
Conclusions: These results provide a framework for the design of future evolutionary studies of gene expression and demonstrate the utility of comparing RNA-seq data across studies.
Figures
References
-
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams BA, Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See L-H, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

