The Kringle-like Domain Facilitates Post-endoplasmic Reticulum Changes to Premelanosome Protein (PMEL) Oligomerization and Disulfide Bond Configuration and Promotes Amyloid Formation
- PMID: 26694611
- PMCID: PMC4751398
- DOI: 10.1074/jbc.M115.692442
The Kringle-like Domain Facilitates Post-endoplasmic Reticulum Changes to Premelanosome Protein (PMEL) Oligomerization and Disulfide Bond Configuration and Promotes Amyloid Formation
Abstract
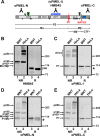
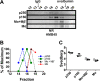
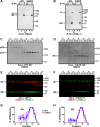
The formation of functional amyloid must be carefully regulated to prevent the accumulation of potentially toxic products. Premelanosome protein (PMEL) forms non-toxic functional amyloid fibrils that assemble into sheets upon which melanins ultimately are deposited within the melanosomes of pigment cells. PMEL is synthesized in the endoplasmic reticulum but forms amyloid only within post-Golgi melanosome precursors; thus, PMEL must traverse the secretory pathway in a non-amyloid form. Here, we identified two pre-amyloid PMEL intermediates that likely regulate the timing of fibril formation. Analyses by non-reducing SDS-PAGE, size exclusion chromatography, and sedimentation velocity revealed two native high Mr disulfide-bonded species that contain Golgi-modified forms of PMEL. These species correspond to disulfide bond-containing dimeric and monomeric PMEL isoforms that contain no other proteins as judged by two-dimensional PAGE of metabolically labeled/immunoprecipitated PMEL and by mass spectrometry of affinity-purified complexes. Metabolic pulse-chase analyses, small molecule inhibitor treatments, and evaluation of site-directed mutants suggest that the PMEL dimer forms around the time of endoplasmic reticulum exit and is resolved by disulfide bond rearrangement into a monomeric form within the late Golgi or a post-Golgi compartment. Mutagenesis of individual cysteine residues within the non-amyloid cysteine-rich Kringle-like domain stabilizes the disulfide-bonded dimer and impairs fibril formation as determined by electron microscopy. Our data show that the Kringle-like domain facilitates the resolution of disulfide-bonded PMEL dimers and promotes PMEL functional amyloid formation, thereby suggesting that PMEL dimers must be resolved to monomers to generate functional amyloid fibrils.
Keywords: Pmel17; cysteine-mediated cross-linking; disulfide; endosome; fibril; melanogenesis; melanosome; membrane trafficking; oxidation-reduction (redox); protein aggregation.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures








References
-
- Chiti F., and Dobson C. M. (2006) Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 - PubMed
-
- Skovronsky D. M., Lee V. M., and Trojanowski J. Q. (2006) Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu. Rev. Pathol. 1, 151–170 - PubMed
-
- Fowler D. M., Koulov A. V., Balch W. E., and Kelly J. W. (2007) Functional amyloid: from bacteria to humans. Trends Biochem. Sci. 32, 217–224 - PubMed
-
- Majumdar A., Cesario W. C., White-Grindley E., Jiang H., Ren F., Khan M. R., Li L., Choi E. M., Kannan K., Guo F., Unruh J., Slaughter B., and Si K. (2012) Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 148, 515–529 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

