Molecular Basis of Enhanced Activity in Factor VIIa-Trypsin Variants Conveys Insights into Tissue Factor-mediated Allosteric Regulation of Factor VIIa Activity
- PMID: 26694616
- PMCID: PMC4813490
- DOI: 10.1074/jbc.M115.698613
Molecular Basis of Enhanced Activity in Factor VIIa-Trypsin Variants Conveys Insights into Tissue Factor-mediated Allosteric Regulation of Factor VIIa Activity
Abstract
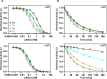
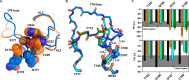
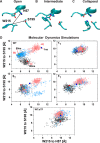
The complex of coagulation factor VIIa (FVIIa), a trypsin-like serine protease, and membrane-bound tissue factor (TF) initiates blood coagulation upon vascular injury. Binding of TF to FVIIa promotes allosteric conformational changes in the FVIIa protease domain and improves its catalytic properties. Extensive studies have revealed two putative pathways for this allosteric communication. Here we provide further details of this allosteric communication by investigating FVIIa loop swap variants containing the 170 loop of trypsin that display TF-independent enhanced activity. Using x-ray crystallography, we show that the introduced 170 loop from trypsin directly interacts with the FVIIa active site, stabilizing segment 215-217 and activation loop 3, leading to enhanced activity. Molecular dynamics simulations and novel fluorescence quenching studies support that segment 215-217 conformation is pivotal to the enhanced activity of the FVIIa variants. We speculate that the allosteric regulation of FVIIa activity by TF binding follows a similar path in conjunction with protease domain N terminus insertion, suggesting a more complete molecular basis of TF-mediated allosteric enhancement of FVIIa activity.
Keywords: allosteric regulation; coagulation factor; molecular dynamics; serine protease; x-ray crystallography.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


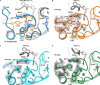

References
-
- Davie E. W., Fujikawa K., and Kisiel W. (1991) The coagulation cascade: initiation, maintenance, and regulation. Biochemistry 30, 10363–10370 - PubMed
-
- Banner D. W., D'Arcy A., Chène C., Winkler F. K., Guha A., Konigsberg W. H., Nemerson Y., and Kirchhofer D. (1996) The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature 380, 41–46 - PubMed
-
- Huber R., and Bode W. (1978) Structural basis of the activation and action of trypsin. Acc. Chem. Res. 266, 114–122
-
- Higashi S., Nishimura H., Aita K., and Iwanaga S. (1994) Identification of regions of bovine factor VII essential for binding to tissue factor. J. Biol. Chem. 269, 18891–18898 - PubMed
-
- Rand K. D., Andersen M. D., Olsen O. H., Jørgensen T. J., Ostergaard H., Jensen O. N., Stennicke H. R., and Persson E. (2008) The origins of enhanced activity in factor VIIa analogs and the interplay between key allosteric sites revealed by hydrogen exchange mass spectrometry. J. Biol. Chem. 283, 13378–13387 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

