PROXIMAL: a method for Prediction of Xenobiotic Metabolism
- PMID: 26695483
- PMCID: PMC4687097
- DOI: 10.1186/s12918-015-0241-4
PROXIMAL: a method for Prediction of Xenobiotic Metabolism
Abstract
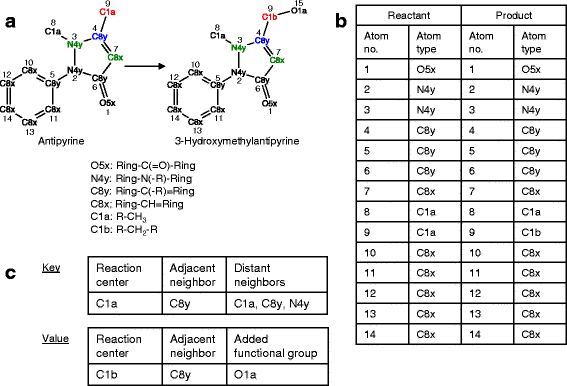
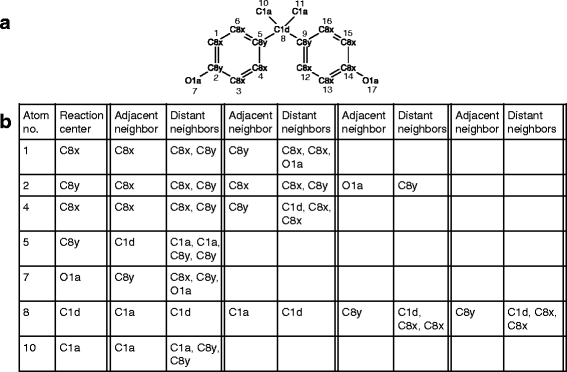
Background: Contamination of the environment with bioactive chemicals has emerged as a potential public health risk. These substances that may cause distress or disease in humans can be found in air, water and food supplies. An open question is whether these chemicals transform into potentially more active or toxic derivatives via xenobiotic metabolizing enzymes expressed in the body. We present a new prediction tool, which we call PROXIMAL (Prediction of Xenobiotic Metabolism) for identifying possible transformation products of xenobiotic chemicals in the liver. Using reaction data from DrugBank and KEGG, PROXIMAL builds look-up tables that catalog the sites and types of structural modifications performed by Phase I and Phase II enzymes. Given a compound of interest, PROXIMAL searches for substructures that match the sites cataloged in the look-up tables, applies the corresponding modifications to generate a panel of possible transformation products, and ranks the products based on the activity and abundance of the enzymes involved.
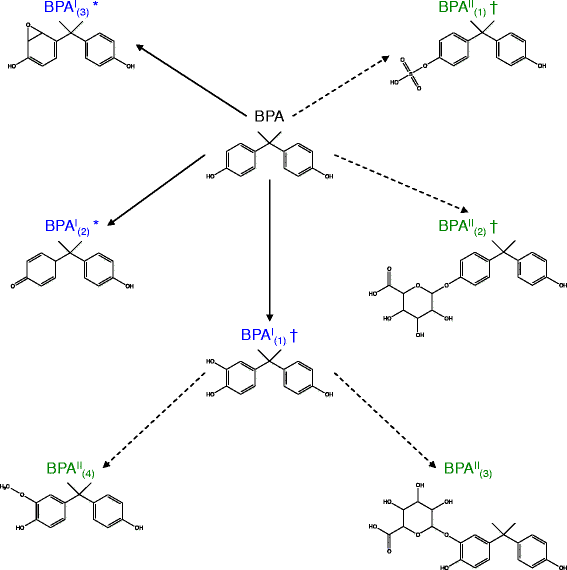
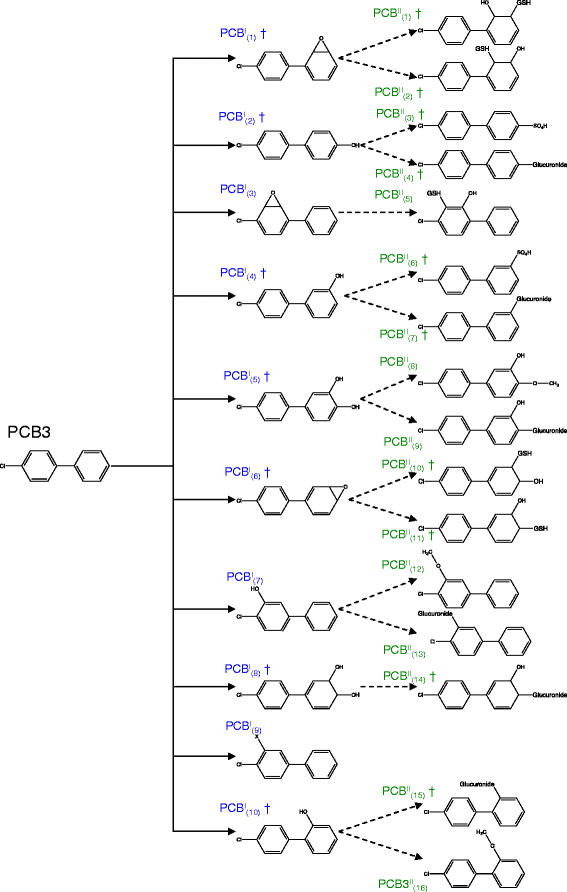
Results: PROXIMAL generates transformations that are specific for the chemical of interest by analyzing the chemical's substructures. We evaluate the accuracy of PROXIMAL's predictions through case studies on two environmental chemicals with suspected endocrine disrupting activity, bisphenol A (BPA) and 4-chlorobiphenyl (PCB3). Comparisons with published reports confirm 5 out of 7 and 17 out of 26 of the predicted derivatives for BPA and PCB3, respectively. We also compare biotransformation predictions generated by PROXIMAL with those generated by METEOR and Metaprint2D-react, two other prediction tools.
Conclusions: PROXIMAL can predict transformations of chemicals that contain substructures recognizable by human liver enzymes. It also has the ability to rank the predicted metabolites based on the activity and abundance of enzymes involved in xenobiotic transformation.
Figures


Similar articles
-
Modulation of metabolizing enzymes by bisphenol a in human and animal models.Chem Res Toxicol. 2014 Sep 15;27(9):1463-73. doi: 10.1021/tx500087p. Epub 2014 Aug 29. Chem Res Toxicol. 2014. PMID: 25142872 Review.
-
Discovery of a widespread metabolic pathway within and among phenolic xenobiotics.Proc Natl Acad Sci U S A. 2017 Jun 6;114(23):6062-6067. doi: 10.1073/pnas.1700558114. Epub 2017 May 23. Proc Natl Acad Sci U S A. 2017. PMID: 28536195 Free PMC article.
-
Nuclear receptor-mediated transcriptional regulation in Phase I, II, and III xenobiotic metabolizing systems.Drug Metab Pharmacokinet. 2006 Dec;21(6):437-57. doi: 10.2133/dmpk.21.437. Drug Metab Pharmacokinet. 2006. PMID: 17220560 Review.
-
Metabolism of xenobiotics of human environments.Prog Mol Biol Transl Sci. 2012;112:31-88. doi: 10.1016/B978-0-12-415813-9.00003-9. Prog Mol Biol Transl Sci. 2012. PMID: 22974737 Review.
-
In situ sites for xenobiotic activation and detoxication: implications for the differential susceptibility of cells to the toxic actions of environmental chemicals.Prog Histochem Cytochem. 1991;23(1-4):32-44. doi: 10.1016/s0079-6336(11)80167-1. Prog Histochem Cytochem. 1991. PMID: 1947157
Cited by
-
Analysis of metabolic network disruption in engineered microbial hosts due to enzyme promiscuity.Metab Eng Commun. 2021 Mar 7;12:e00170. doi: 10.1016/j.mec.2021.e00170. eCollection 2021 Jun. Metab Eng Commun. 2021. PMID: 33850714 Free PMC article.
-
Constructing xenobiotic maps of metabolism to predict enzymes catalyzing metabolites capable of binding to DNA.BMC Bioinformatics. 2021 Sep 21;22(1):450. doi: 10.1186/s12859-021-04363-6. BMC Bioinformatics. 2021. PMID: 34548010 Free PMC article.
-
Extending PROXIMAL to predict degradation pathways of phenolic compounds in the human gut microbiota.NPJ Syst Biol Appl. 2024 May 27;10(1):56. doi: 10.1038/s41540-024-00381-1. NPJ Syst Biol Appl. 2024. PMID: 38802371 Free PMC article.
-
Pickaxe: a Python library for the prediction of novel metabolic reactions.BMC Bioinformatics. 2023 Mar 22;24(1):106. doi: 10.1186/s12859-023-05149-8. BMC Bioinformatics. 2023. PMID: 36949401 Free PMC article.
-
System Prediction and Validation of TCM for Chronic Myeloid Leukemia Treatment from the Perspective of Low-Toxicity Chemotherapy: A Stilbene α-Viniferin Has a Proapoptotic Effect on K562 Cells via the Mitochondrial Pathway.Evid Based Complement Alternat Med. 2020 Feb 10;2020:1986962. doi: 10.1155/2020/1986962. eCollection 2020. Evid Based Complement Alternat Med. 2020. PMID: 32104190 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

