Diversity and Habitat Preferences of Cultivated and Uncultivated Aerobic Methanotrophic Bacteria Evaluated Based on pmoA as Molecular Marker
- PMID: 26696968
- PMCID: PMC4678205
- DOI: 10.3389/fmicb.2015.01346
Diversity and Habitat Preferences of Cultivated and Uncultivated Aerobic Methanotrophic Bacteria Evaluated Based on pmoA as Molecular Marker
Abstract
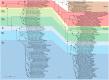
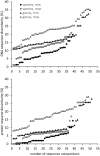
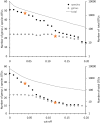
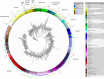
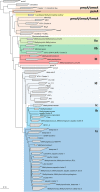
Methane-oxidizing bacteria are characterized by their capability to grow on methane as sole source of carbon and energy. Cultivation-dependent and -independent methods have revealed that this functional guild of bacteria comprises a substantial diversity of organisms. In particular the use of cultivation-independent methods targeting a subunit of the particulate methane monooxygenase (pmoA) as functional marker for the detection of aerobic methanotrophs has resulted in thousands of sequences representing "unknown methanotrophic bacteria." This limits data interpretation due to restricted information about these uncultured methanotrophs. A few groups of uncultivated methanotrophs are assumed to play important roles in methane oxidation in specific habitats, while the biology behind other sequence clusters remains still largely unknown. The discovery of evolutionary related monooxygenases in non-methanotrophic bacteria and of pmoA paralogs in methanotrophs requires that sequence clusters of uncultivated organisms have to be interpreted with care. This review article describes the present diversity of cultivated and uncultivated aerobic methanotrophic bacteria based on pmoA gene sequence diversity. It summarizes current knowledge about cultivated and major clusters of uncultivated methanotrophic bacteria and evaluates habitat specificity of these bacteria at different levels of taxonomic resolution. Habitat specificity exists for diverse lineages and at different taxonomic levels. Methanotrophic genera such as Methylocystis and Methylocaldum are identified as generalists, but they harbor habitat specific methanotrophs at species level. This finding implies that future studies should consider these diverging preferences at different taxonomic levels when analyzing methanotrophic communities.
Keywords: diversity; ecological niche; habitat specificity; methanotrophic bacteria; phylogeny; pmoA.
Figures


Similar articles
-
Diversity of the particulate methane monooxygenase gene in methanotrophic samples from different rice field soils in China and the Philippines.Syst Appl Microbiol. 2002 Aug;25(2):267-74. doi: 10.1078/0723-2020-00104. Syst Appl Microbiol. 2002. PMID: 12353882
-
The active methanotrophic community in hydromorphic soils changes in response to changing methane concentration.Environ Microbiol. 2006 Feb;8(2):321-33. doi: 10.1111/j.1462-2920.2005.00898.x. Environ Microbiol. 2006. PMID: 16423018
-
A pmoA-based study reveals dominance of yet uncultured Type I methanotrophs in rhizospheres of an organically fertilized rice field in India.3 Biotech. 2016 Dec;6(2):135. doi: 10.1007/s13205-016-0453-3. Epub 2016 Jun 16. 3 Biotech. 2016. PMID: 28330207 Free PMC article.
-
Methanotrophic bacteria.Microbiol Rev. 1996 Jun;60(2):439-71. doi: 10.1128/mr.60.2.439-471.1996. Microbiol Rev. 1996. PMID: 8801441 Free PMC article. Review.
-
Recent findings in methanotrophs: genetics, molecular ecology, and biopotential.Appl Microbiol Biotechnol. 2024 Dec;108(1):60. doi: 10.1007/s00253-023-12978-3. Epub 2024 Jan 6. Appl Microbiol Biotechnol. 2024. PMID: 38183483 Review.
Cited by
-
Aerobic and anaerobic methane oxidation in a seasonally anoxic basin.Limnol Oceanogr. 2022 Jun;67(6):1257-1273. doi: 10.1002/lno.12074. Epub 2022 Mar 28. Limnol Oceanogr. 2022. PMID: 36248250 Free PMC article.
-
Methylococcaceae are the dominant active aerobic methanotrophs in a Chinese tidal marsh.Environ Sci Pollut Res Int. 2019 Jan;26(1):636-646. doi: 10.1007/s11356-018-3560-3. Epub 2018 Nov 8. Environ Sci Pollut Res Int. 2019. PMID: 30411293
-
Methylotetracoccus oryzae Strain C50C1 Is a Novel Type Ib Gammaproteobacterial Methanotroph Adapted to Freshwater Environments.mSphere. 2019 Jun 5;4(3):e00631-18. doi: 10.1128/mSphere.00631-18. mSphere. 2019. PMID: 31167950 Free PMC article.
-
Complete Genome Sequence of the Aerobic Facultative Methanotroph Methylocella tundrae Strain T4.Microbiol Resour Announc. 2019 May 16;8(20):e00286-19. doi: 10.1128/MRA.00286-19. Microbiol Resour Announc. 2019. PMID: 31097502 Free PMC article.
-
Long-Term Rewetting of Three Formerly Drained Peatlands Drives Congruent Compositional Changes in Pro- and Eukaryotic Soil Microbiomes through Environmental Filtering.Microorganisms. 2020 Apr 10;8(4):550. doi: 10.3390/microorganisms8040550. Microorganisms. 2020. PMID: 32290343 Free PMC article.
References
-
- Amaral J. A., Archambault C., Richards S. R., Knowles R. (1995). Denitrification associated with Groups I and II methanotrophs in a gradient enrichment system. FEMS Microbiol. Ecol. 18, 289–298. 10.1111/j.1574-6941.1995.tb00185.x - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

