Comprehensive Characterization of Escherichia coli O104:H4 Isolated from Patients in the Netherlands
- PMID: 26696970
- PMCID: PMC4667096
- DOI: 10.3389/fmicb.2015.01348
Comprehensive Characterization of Escherichia coli O104:H4 Isolated from Patients in the Netherlands
Abstract
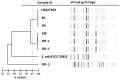
In 2011, a Shiga toxin-producing Enteroaggregative Escherichia coli (EAEC Stx2a+) O104:H4 strain caused a serious outbreak of acute gastroenteritis and hemolytic-uremic syndrome (HUS) in Germany. In 2013, E. coli O104:H4 isolates were obtained from a patient with HUS and her friend showing only gastrointestinal complaints. The antimicrobial resistance and virulence profiles of these isolates together with three EAEC Stx2a+ O104:H4 isolates from 2011 were determined and compared. Whole-genome sequencing (WGS) was performed for detailed characterization and to determine genetic relationship of the isolates. Four additional genomes of EAEC Stx2a+ O104:H4 isolates of 2009 and 2011 available on NCBI were included in the virulence and phylogenetic analysis. All E. coli O104:H4 isolates tested were positive for stx2a, aatA, and terD but were negative for escV. All, except one 2011 isolate, were positive for aggR and were therefore considered EAEC. The EAEC Stx2a+ O104:H4 isolates of 2013 belonged to sequence type (ST) ST678 as the 2011 isolates and showed slightly different resistance and virulence patterns compared to the 2011 isolates. Core-genome phylogenetic analysis showed that the isolates of 2013 formed a separate cluster from the isolates of 2011 and 2009 by 27 and 20 different alleles, respectively. In addition, only a one-allele difference was found between the isolate of the HUS-patient and that of her friend. Our study shows that EAEC Stx2a+ O104:H4 strains highly similar to the 2011 outbreak clone in their core genome are still circulating necessitating proper surveillance to prevent further outbreaks with these potentially pathogenic strains. In addition, WGS not only provided a detailed characterization of the isolates but its high discriminatory power also enabled us to discriminate the 2013 isolates from the isolates of 2009 and 2011 expediting the use of WGS in public health services to rapidly apply proper infection control strategies.
Keywords: Shiga toxin-producing E. coli –STEC; antimicrobial resistance; enterohemorrhagic E. coli –EHEC; hemolytic uremic syndrome; outbreak; phylogenetic analysis; whole genome sequencing.
Figures
Similar articles
-
Shiga toxin-producing Escherichia coli strains from cattle as a source of the Stx2a bacteriophages present in enteroaggregative Escherichia coli O104:H4 strains.Int J Med Microbiol. 2013 Dec;303(8):595-602. doi: 10.1016/j.ijmm.2013.08.001. Epub 2013 Aug 16. Int J Med Microbiol. 2013. PMID: 24012149
-
Escherichia coli O104:H4 Pathogenesis: an Enteroaggregative E. coli/Shiga Toxin-Producing E. coli Explosive Cocktail of High Virulence.Microbiol Spectr. 2014 Dec;2(6). doi: 10.1128/microbiolspec.EHEC-0008-2013. Microbiol Spectr. 2014. PMID: 26104460 Review.
-
No evidence of the Shiga toxin-producing E. coli O104:H4 outbreak strain or enteroaggregative E. coli (EAEC) found in cattle faeces in northern Germany, the hotspot of the 2011 HUS outbreak area.Gut Pathog. 2011 Nov 3;3(1):17. doi: 10.1186/1757-4749-3-17. Gut Pathog. 2011. PMID: 22051440 Free PMC article.
-
Fitness of Enterohemorrhagic Escherichia coli (EHEC)/Enteroaggregative E. coli O104:H4 in Comparison to That of EHEC O157: Survival Studies in Food and In Vitro.Appl Environ Microbiol. 2016 Oct 14;82(21):6326-6334. doi: 10.1128/AEM.01796-16. Print 2016 Nov 1. Appl Environ Microbiol. 2016. PMID: 27542931 Free PMC article.
-
Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain.Int Microbiol. 2011 Sep;14(3):121-41. doi: 10.2436/20.1501.01.142. Int Microbiol. 2011. PMID: 22101411 Review.
Cited by
-
WGS of intrauterine E. coli from cows with early postpartum uterine infection reveals a non-uterine specific genotype and virulence factors.mBio. 2024 Jun 12;15(6):e0102724. doi: 10.1128/mbio.01027-24. Epub 2024 May 14. mBio. 2024. PMID: 38742889 Free PMC article.
-
Shiga Toxin-Producing and Enteroaggregative Escherichia coli in Animal, Foods, and Humans: Pathogenicity Mechanisms, Detection Methods, and Epidemiology.Curr Microbiol. 2020 Apr;77(4):612-620. doi: 10.1007/s00284-019-01842-1. Epub 2019 Dec 13. Curr Microbiol. 2020. PMID: 31834432 Review.
-
Whole genome sequencing reveals high clonal diversity of Escherichia coli isolated from patients in a tertiary care hospital in Moshi, Tanzania.Antimicrob Resist Infect Control. 2018 Jun 8;7:72. doi: 10.1186/s13756-018-0361-x. eCollection 2018. Antimicrob Resist Infect Control. 2018. PMID: 29977533 Free PMC article.
-
Diversity of the Tellurite Resistance Gene Operon in Escherichia coli.Front Microbiol. 2021 May 28;12:681175. doi: 10.3389/fmicb.2021.681175. eCollection 2021. Front Microbiol. 2021. PMID: 34122392 Free PMC article.
-
Sporadic Occurrence of Enteroaggregative Shiga Toxin-Producing Escherichia coli O104:H4 Similar to 2011 Outbreak Strain.Emerg Infect Dis. 2022 Sep;28(9):1890-1894. doi: 10.3201/eid2809.220037. Emerg Infect Dis. 2022. PMID: 35997633 Free PMC article.
References
-
- Ahmed S. A., Awosika J., Baldwin C., Bishop-Lilly K. A., Biswas B., Broomall S., et al. (2012). Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including Shiga toxin encoding phage stx2. PLoS ONE 7:e48228 10.1371/journal.pone.0048228 - DOI - PMC - PubMed
-
- Bielaszewska M., Mellmann A., Zhang W., Köck R., Fruth A., Bauwens A., et al. (2011). Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11 671–676. 10.1016/S1473-3099(11)70165-7 - DOI - PubMed
-
- Brzuszkiewicz E., Thürmer A., Schuldes J., Leimbach A., Liesegang H., Meyer F. D., et al. (2011). Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: entero-aggregative-haemorrhagic Escherichia coli (EAHEC). Arch. Microbiol. 193 883–891. 10.1007/s00203-011-0725-6 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

