Literature Mining and Ontology based Analysis of Host-Brucella Gene-Gene Interaction Network
- PMID: 26696993
- PMCID: PMC4673313
- DOI: 10.3389/fmicb.2015.01386
Literature Mining and Ontology based Analysis of Host-Brucella Gene-Gene Interaction Network
Abstract
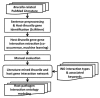
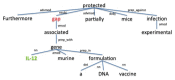
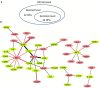
Brucella is an intracellular bacterium that causes chronic brucellosis in humans and various mammals. The identification of host-Brucella interaction is crucial to understand host immunity against Brucella infection and Brucella pathogenesis against host immune responses. Most of the information about the inter-species interactions between host and Brucella genes is only available in the text of the scientific publications. Many text-mining systems for extracting gene and protein interactions have been proposed. However, only a few of them have been designed by considering the peculiarities of host-pathogen interactions. In this paper, we used a text mining approach for extracting host-Brucella gene-gene interactions from the abstracts of articles in PubMed. The gene-gene interactions here represent the interactions between genes and/or gene products (e.g., proteins). The SciMiner tool, originally designed for detecting mammalian gene/protein names in text, was extended to identify host and Brucella gene/protein names in the abstracts. Next, sentence-level and abstract-level co-occurrence based approaches, as well as sentence-level machine learning based methods, originally designed for extracting intra-species gene interactions, were utilized to extract the interactions among the identified host and Brucella genes. The extracted interactions were manually evaluated. A total of 46 host-Brucella gene interactions were identified and represented as an interaction network. Twenty four of these interactions were identified from sentence-level processing. Twenty two additional interactions were identified when abstract-level processing was performed. The Interaction Network Ontology (INO) was used to represent the identified interaction types at a hierarchical ontology structure. Ontological modeling of specific gene-gene interactions demonstrates that host-pathogen gene-gene interactions occur at experimental conditions which can be ontologically represented. Our results show that the introduced literature mining and ontology-based modeling approach are effective in retrieving and analyzing host-pathogen gene-gene interaction networks.
Keywords: Brucella; Interaction Network Ontology (INO); SciMiner; host and pathogen gene name recognition; host–pathogen interaction extraction; support vector machines (SVM); text mining.
Figures



References
-
- Al-Mariri A., Tibor A., Mertens P., De Bolle X., Michel P., Godefroid J., et al. (2001). Protection of BALB/c mice against Brucella abortus 544 challenge by vaccination with bacterioferritin or P39 recombinant proteins with CpG oligodeoxynucleotides as adjuvant. Infect. Immun. 69 4816–4822. 10.1128/IAI.69.8.4816-4822.2001 - DOI - PMC - PubMed
-
- Blaschke C., Valencia A. (2002). The frame-based module of the SUISEKI information extraction system. IEEE Intell. Syst. 17 14–20. 10.1109/MIS.2002.999215 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

