Novel Routes for Improving Biocontrol Activity of Bacillus Based Bioinoculants
- PMID: 26696998
- PMCID: PMC4674565
- DOI: 10.3389/fmicb.2015.01395
Novel Routes for Improving Biocontrol Activity of Bacillus Based Bioinoculants
Abstract
Biocontrol (BC) formulations prepared from plant-growth-promoting bacteria are increasingly applied in sustainable agriculture. Especially inoculants prepared from endospore-forming Bacillus strains have been proven as efficient and environmental-friendly alternative to chemical pesticides due to their long shelf life, which is comparable with that of agrochemicals. However, these formulations of the first generation are sometimes hampered in their action and do not fulfill in each case the expectations of the appliers. In this review we use the well-known plant-associated Bacillus amyloliquefaciens type strain FZB42 as example for the successful application of different techniques offered today by comparative, evolutionary and functional genomics, site-directed mutagenesis and strain construction including marker removal, for paving the way for preparing a novel generation of BC agents.
Keywords: Bacillus amyloliquefaciens subsp. plantarum; bacillomycin D; bacilysin; biocontrol; harpin genes; mersacidin; plant growth-promotion; surfactin.
Figures
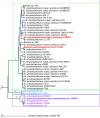
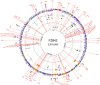
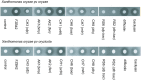

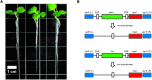
References
-
- Borriss R. (2011). “Use of plant-associated Bacillus strains as biofertilizers and biocontrol agents,” in Bacteria in Agrobiology: Plant Growth Responses, ed. Maheshwari D. K. (Berlin: Springer; ), 41–76.
-
- Borriss R. (2013). “Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42,” in Molecular Microbial Ecology of the Rhizosphere, ed. de Bruijn F. J. (Hoboken, NJ: Wiley Blackwell; ), 883–898.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

