Comprehensive Expression Profiling of Rice Tetraspanin Genes Reveals Diverse Roles During Development and Abiotic Stress
- PMID: 26697042
- PMCID: PMC4675852
- DOI: 10.3389/fpls.2015.01088
Comprehensive Expression Profiling of Rice Tetraspanin Genes Reveals Diverse Roles During Development and Abiotic Stress
Abstract
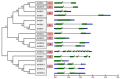
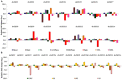
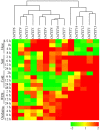
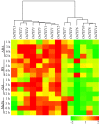
Tetraspanin family is comprised of evolutionarily conserved integral membrane proteins. The incredible ability of tetraspanins to form 'micro domain complexes' and their preferential targeting to membranes emphasizes their active association with signal recognition and communication with neighboring cells, thus acting as key modulators of signaling cascades. In animals, tetraspanins are associated with multitude of cellular processes. Unlike animals, the biological relevance of tetraspanins in plants has not been well investigated. In Arabidopsis tetraspanins are known to contribute in important plant development processes such as leaf morphogenesis, root, and floral organ formation. In the present study we investigated the genomic organization, chromosomal distribution, phylogeny and domain structure of 15 rice tetraspanin proteins (OsTETs). OsTET proteins had similar domain structure and signature 'GCCK/R' motif as reported in Arabidopsis. Comprehensive expression profiling of OsTET genes suggested their possible involvement during rice development. While OsTET9 and 10 accumulated predominantly in flowers, OsTET5, 8, and 12 were preferentially expressed in root tissues. Noticeably, seven OsTETs exhibited more than twofold up regulation at early stages of flag leaf senescence in rice. Furthermore, several OsTETs were differentially regulated in rice seedlings exposed to abiotic stresses, exogenous treatment of hormones and nutrient deprivation. Transient subcellular localization studies of eight OsTET proteins in tobacco epidermal cells showed that these proteins localized in plasma membrane. The present study provides valuable insights into the possible roles of tetraspanins in regulating development and defining response to abiotic stresses in rice. Targeted proteomic studies would be useful in identification of their interacting partners under different conditions and ultimately their biological function in plants.
Keywords: abiotic stress; gene expression; hormone; nutrient deprivation; rice; tetraspanin.
Figures




Similar articles
-
Differential tetraspanin genes expression and subcellular localization during mutualistic interactions in Phaseolus vulgaris.PLoS One. 2019 Aug 22;14(8):e0219765. doi: 10.1371/journal.pone.0219765. eCollection 2019. PLoS One. 2019. PMID: 31437164 Free PMC article.
-
Rice tetraspanins express in specific domains of diverse tissues and regulate plant architecture and root growth.Plant J. 2024 Feb;117(3):892-908. doi: 10.1111/tpj.16536. Epub 2023 Nov 13. Plant J. 2024. PMID: 37955978
-
Arabidopsis Tetraspanins Facilitate Virus Infection via Membrane-Recognition GCCK/RP Motif and Cysteine Residues.Front Plant Sci. 2022 Mar 3;13:805633. doi: 10.3389/fpls.2022.805633. eCollection 2022. Front Plant Sci. 2022. PMID: 35310653 Free PMC article.
-
Tetraspanin genes in plants.Plant Sci. 2012 Jul;190:9-15. doi: 10.1016/j.plantsci.2012.03.005. Epub 2012 Mar 23. Plant Sci. 2012. PMID: 22608515 Review.
-
TETRASPANINs in Plants.Front Plant Sci. 2017 Apr 18;8:545. doi: 10.3389/fpls.2017.00545. eCollection 2017. Front Plant Sci. 2017. PMID: 28458676 Free PMC article. Review.
Cited by
-
Fine mapping and identification of two NtTOM2A homeologs responsible for tobacco mosaic virus replication in tobacco (Nicotiana tabacum L.).BMC Plant Biol. 2024 Jan 24;24(1):67. doi: 10.1186/s12870-024-04744-y. BMC Plant Biol. 2024. PMID: 38262958 Free PMC article.
-
Genome-Wide Association Mapping of Salinity Tolerance at the Seedling Stage in a Panel of Vietnamese Landraces Reveals New Valuable QTLs for Salinity Stress Tolerance Breeding in Rice.Plants (Basel). 2021 May 28;10(6):1088. doi: 10.3390/plants10061088. Plants (Basel). 2021. PMID: 34071570 Free PMC article.
-
Differential tetraspanin genes expression and subcellular localization during mutualistic interactions in Phaseolus vulgaris.PLoS One. 2019 Aug 22;14(8):e0219765. doi: 10.1371/journal.pone.0219765. eCollection 2019. PLoS One. 2019. PMID: 31437164 Free PMC article.
-
Genome-wide identification, characterization, and expression analysis unveil the roles of pseudouridine synthase (PUS) family proteins in rice development and stress response.Physiol Mol Biol Plants. 2023 Dec;29(12):1981-2004. doi: 10.1007/s12298-023-01396-4. Epub 2023 Nov 30. Physiol Mol Biol Plants. 2023. PMID: 38222285 Free PMC article.
-
Advanced High-Throughput Root Phenotyping and GWAS Identifies Key Genomic Regions in Cowpea During Vegetative Growth Stage.Physiol Plant. 2025 Jul-Aug;177(4):e70375. doi: 10.1111/ppl.70375. Physiol Plant. 2025. PMID: 40611537 Free PMC article.
References
-
- Berditchevski F. (2001). Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114 4143–4151. - PubMed
-
- Boavida L. C., Qin P., Broz M., Becker J. D., Mccormick S. (2013). Arabidopsis tetraspanins are confined to discrete expression domains and cell types in reproductive tissues and form homo– and heterodimers when expressed in yeast. Plant Physiol. 163 696–712. 10.1104/pp.113.216598 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

