Flower Development and Perianth Identity Candidate Genes in the Basal Angiosperm Aristolochia fimbriata (Piperales: Aristolochiaceae)
- PMID: 26697047
- PMCID: PMC4675851
- DOI: 10.3389/fpls.2015.01095
Flower Development and Perianth Identity Candidate Genes in the Basal Angiosperm Aristolochia fimbriata (Piperales: Aristolochiaceae)
Abstract
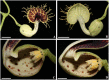
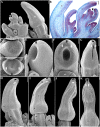
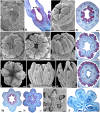
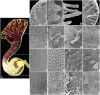
Aristolochia fimbriata (Aristolochiaceae: Piperales) exhibits highly synorganized flowers with a single convoluted structure forming a petaloid perianth that surrounds the gynostemium, putatively formed by the congenital fusion between stamens and the upper portion of the carpels. Here we present the flower development and morphology of A. fimbriata, together with the expression of the key regulatory genes that participate in flower development, particularly those likely controlling perianth identity. A. fimbriata is a member of the magnoliids, and thus gene expression detected for all ABCE MADS-box genes in this taxon, can also help to elucidate patterns of gene expression prior the independent duplications of these genes in eudicots and monocots. Using both floral development and anatomy in combination with the isolation of MADS-box gene homologs, gene phylogenetic analyses and expression studies (both by reverse transcription PCR and in situ hybridization), we present hypotheses on floral organ identity genes involved in the formation of this bizarre flower. We found that most MADS-box genes were expressed in vegetative and reproductive tissues with the exception of AfimSEP2, AfimAGL6, and AfimSTK transcripts that are only found in flowers and capsules but are not detected in leaves. Two genes show ubiquitous expression; AfimFUL that is found in all floral organs at all developmental stages as well as in leaves and capsules, and AfimAG that has low expression in leaves and is found in all floral organs at all stages with a considerable reduction of expression in the limb of anthetic flowers. Our results indicate that expression of AfimFUL is indicative of pleiotropic roles and not of a perianth identity specific function. On the other hand, expression of B-class genes, AfimAP3 and AfimPI, suggests their conserved role in stamen identity and corroborates that the perianth is sepal and not petal-derived. Our data also postulates an AGL6 ortholog as a candidate gene for sepal identity in the Aristolochiaceae and provides testable hypothesis for a modified ABCE model in synorganized magnoliid flowers.
Keywords: AGAMOUS-like6; APETALA3; Aristolochia fimbriata; FRUITFULL; MADS-box genes; PISTILLATA; magnoliids; perianth.
Figures



Similar articles
-
Deep into the Aristolochia Flower: Expression of C, D, and E-Class Genes in Aristolochia fimbriata (Aristolochiaceae).J Exp Zool B Mol Dev Evol. 2017 Jan;328(1-2):55-71. doi: 10.1002/jez.b.22686. Epub 2016 Aug 10. J Exp Zool B Mol Dev Evol. 2017. PMID: 27507740
-
Floral MADS-box protein interactions in the early diverging angiosperm Aristolochia fimbriata Cham. (Aristolochiaceae: Piperales).Evol Dev. 2019 Mar;21(2):96-110. doi: 10.1111/ede.12282. Epub 2019 Feb 8. Evol Dev. 2019. PMID: 30734997
-
APETALA3 and PISTILLATA homologs exhibit novel expression patterns in the unique perianth of Aristolochia (Aristolochiaceae).Evol Dev. 2004 Nov-Dec;6(6):449-58. doi: 10.1111/j.1525-142X.2004.04053.x. Evol Dev. 2004. PMID: 15509227
-
How to Evolve a Perianth: A Review of Cadastral Mechanisms for Perianth Identity.Front Plant Sci. 2018 Oct 29;9:1573. doi: 10.3389/fpls.2018.01573. eCollection 2018. Front Plant Sci. 2018. PMID: 30420867 Free PMC article. Review.
-
To B or Not to B a flower: the role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms.J Hered. 2005 May-Jun;96(3):225-40. doi: 10.1093/jhered/esi033. Epub 2005 Feb 4. J Hered. 2005. PMID: 15695551 Review.
Cited by
-
Expression and Functional Analyses of Nymphaea caerulea MADS-Box Genes Contribute to Clarify the Complex Flower Patterning of Water Lilies.Front Plant Sci. 2021 Sep 22;12:730270. doi: 10.3389/fpls.2021.730270. eCollection 2021. Front Plant Sci. 2021. PMID: 34630477 Free PMC article.
-
Uncovering developmental diversity in the field.Development. 2024 Oct 15;151(20):dev203084. doi: 10.1242/dev.203084. Epub 2024 Aug 19. Development. 2024. PMID: 39158021 Free PMC article.
-
Evolution of major flowering pathway integrators in Orchidaceae.Plant Reprod. 2024 Jun;37(2):85-109. doi: 10.1007/s00497-023-00482-7. Epub 2023 Oct 12. Plant Reprod. 2024. PMID: 37823912 Free PMC article.
-
Model Species to Investigate the Origin of Flowers.Methods Mol Biol. 2023;2686:83-109. doi: 10.1007/978-1-0716-3299-4_4. Methods Mol Biol. 2023. PMID: 37540355
-
The Evolution of euAPETALA2 Genes in Vascular Plants: From Plesiomorphic Roles in Sporangia to Acquired Functions in Ovules and Fruits.Mol Biol Evol. 2021 May 19;38(6):2319-2336. doi: 10.1093/molbev/msab027. Mol Biol Evol. 2021. PMID: 33528546 Free PMC article.
References
-
- Akaike H. (1974). A new look at the statistical model identification. IEEE Trans. Automatic Control 19 716–723. 10.1109/TAC.1974.1100705 - DOI
-
- Alvarez J., Smyth D. R. (2002). CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int. J. Plant Sci. 163 17–41. 10.1086/324178 - DOI
-
- Alvarez Buylla E. R., Ambrose B. A., Flores-Sandoval E., Englund M., Garay-Arroyo A., García-Ponce B., et al. (2010). B-function expression in the flower center underlies the homeotic phenotype of Lacandonia schismatica (Triuridaceae). Plant Cell 22 3543–3559. 10.1105/tpc.109.069153 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

