Unsupervised image segmentation for microarray spots with irregular contours and inner holes
- PMID: 26698293
- PMCID: PMC4690322
- DOI: 10.1186/s12859-015-0842-3
Unsupervised image segmentation for microarray spots with irregular contours and inner holes
Abstract
Background: Microarray analysis represents a powerful way to test scientific hypotheses on the functionality of cells. The measurements consider the whole genome, and the large number of generated data requires sophisticated analysis. To date, no gold-standard for the analysis of microarray images has been established. Due to the lack of a standard approach there is a strong need to identify new processing algorithms.
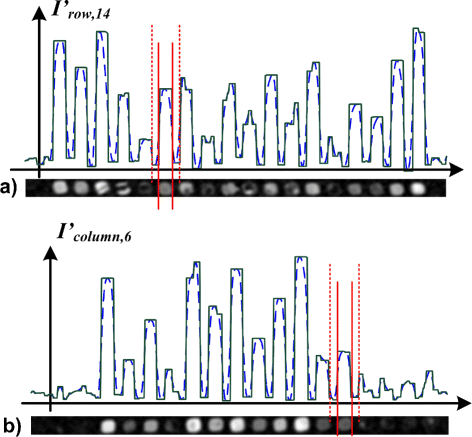
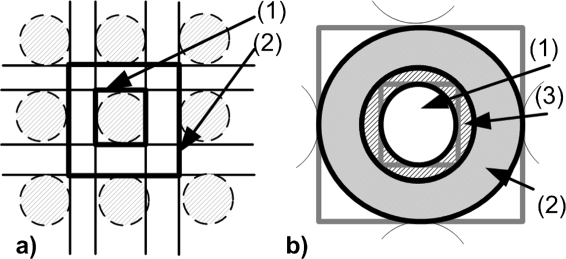
Methods: We propose a novel approach based on hyperbolic partial differential equations (PDEs) for unsupervised spot segmentation. Prior to segmentation, morphological operations were applied for the identification of co-localized groups of spots. A grid alignment was performed to determine the borderlines between rows and columns of spots. PDEs were applied to detect the inflection points within each column and row; vertical and horizontal luminance profiles were evolved respectively. The inflection points of the profiles determined borderlines that confined a spot within adapted rectangular areas. A subsequent k-means clustering determined the pixels of each individual spot and its local background.
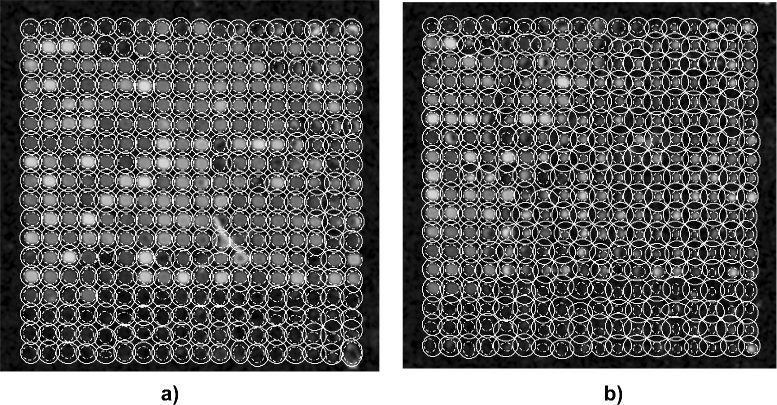
Results: We evaluated the approach for a data set of microarray images taken from the Stanford Microarray Database (SMD). The data set is based on two studies on global gene expression profiles of Arabidopsis Thaliana. We computed values for spot intensity, regression ratio, and coefficient of determination. For spots with irregular contours and inner holes, we found intensity values that were significantly different from those determined by the GenePix Pro microarray analysis software. We determined the set of differentially expressed genes from our intensities and identified more activated genes than were predicted by the GenePix software.
Conclusions: Our method represents a worthwhile alternative and complement to standard approaches used in industry and academy. We highlight the importance of our spot segmentation approach, which identified supplementary important genes, to better explains the molecular mechanisms that are activated in a defense responses to virus and pathogen infection.
Figures




References
-
- Schena M. Microarray Analysis. New York: Wiley; 2003.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

