Esophageal Squamous Cell Carcinoma Cells Modulate Chemokine Expression and Hyaluronan Synthesis in Fibroblasts
- PMID: 26699196
- PMCID: PMC4759185
- DOI: 10.1074/jbc.M115.708909
Esophageal Squamous Cell Carcinoma Cells Modulate Chemokine Expression and Hyaluronan Synthesis in Fibroblasts
Abstract
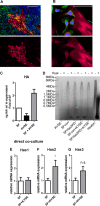
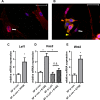
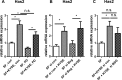
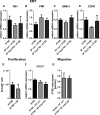
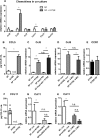
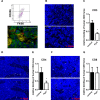
The aim of this study was to characterize the interaction of KYSE-410, an esophageal squamous cell carcinoma cell line, and fibroblasts with respect to the extracellular matrix component hyaluronan (HA) and chemokine expression. KYSE-410 cells induced the mRNA expression of HA synthase 2 (Has2) in normal skin fibroblasts (SF) only in direct co-cultures. Parallel to Has2 mRNA, Has2 antisense RNA (Has2os2) was up-regulated in co-cultures. Knockdown of LEF1, a downstream target of Wnt signaling, abrogated Has2 and Has2os2 induction. After knockdown of Has2 in SF, significantly less α-smooth muscle actin expression was detected in co-cultures. Moreover, it was investigated whether the phenotype of KYSE-410 was affected in co-culture with SF and whether Has2 knockdown in SF had an impact on KYSE-410 cells in co-culture. However, no effects on epithelial-mesenchymal transition markers, proliferation, and migration were detected. In addition to Has2 mRNA, the chemokine CCL5 was up-regulated and CCL11 was down-regulated in SF in co-culture. Furthermore, co-cultures of KYSE-410 cells and cancer-associated fibroblasts (CAF) were investigated. Similar to SF, Has2 and Ccl5 were up-regulated and Ccl11 was down-regulated in CAF in co-culture. Importantly and in contrast to SF, inhibiting HA synthesis by 4-methylumbelliferone abrogated the effect of co-culture on Ccl5 in CAF. Moreover, HA was found to promote adhesion of CD4(+) but not CD8(+) cells to xenogaft tumor tissues. In conclusion, direct co-culture of esophageal squamous cell carcinoma and fibroblasts induced stromal HA synthesis via Wnt/LEF1 and altered the chemokine profile of stromal fibroblasts, which in turn may affect the tumor immune response.
Keywords: CCL11; CCL5; cancer; cell-cell interaction; chemokine; fibroblast; hyaluronan.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Servais C., and Erez N. (2013) From sentinel cells to inflammatory culprits: cancer-associated fibroblasts in tumour-related inflammation. J. Pathol 229, 198–207 - PubMed
-
- Harper J., and Sainson R. C. (2014) Regulation of the anti-tumour immune response by cancer-associated fibroblasts. Semin. Cancer Biol. 25, 69–77 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

