Antigenic and Cryo-Electron Microscopy Structure Analysis of a Chimeric Sapovirus Capsid
- PMID: 26699644
- PMCID: PMC4810723
- DOI: 10.1128/JVI.02916-15
Antigenic and Cryo-Electron Microscopy Structure Analysis of a Chimeric Sapovirus Capsid
Abstract
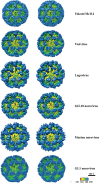
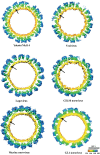
The capsid protein (VP1) of all caliciviruses forms an icosahedral particle with two principal domains, shell (S) and protruding (P) domains, which are connected via a flexible hinge region. The S domain forms a scaffold surrounding the nucleic acid, while the P domains form a homodimer that interacts with receptors. The P domain is further subdivided into two subdomains, termed P1 and P2. The P2 subdomain is likely an insertion in the P1 subdomain; consequently, the P domain is divided into the P1-1, P2, and P1-2 subdomains. In order to investigate capsid antigenicity, N-terminal (N-term)/S/P1-1 and P2/P1-2 were switched between two sapovirus genotypes GI.1 and GI.5. The chimeric VP1 constructs were expressed in insect cells and were shown to self-assemble into virus-like particles (VLPs) morphologically similar to the parental VLPs. Interestingly, the chimeric VLPs had higher levels of cross-reactivities to heterogeneous antisera than the parental VLPs. In order to better understand the antigenicity from a structural perspective, we determined an intermediate-resolution (8.5-Å) cryo-electron microscopy (cryo-EM) structure of a chimeric VLP and developed a VP1 homology model. The cryo-EM structure revealed that the P domain dimers were raised slightly (∼5 Å) above the S domain. The VP1 homology model allowed us predict the S domain (67-229) and P1-1 (229-280), P2 (281-447), and P1-2 (448-567) subdomains. Our results suggested that the raised P dimers might expose immunoreactive S/P1-1 subdomain epitopes. Consequently, the higher levels of cross-reactivities with the chimeric VLPs resulted from a combination of GI.1 and GI.5 epitopes.
Importance: We developed sapovirus chimeric VP1 constructs and produced the chimeric VLPs in insect cells. We found that both chimeric VLPs had a higher level of cross-reactivity against heterogeneous VLP antisera than the parental VLPs. The cryo-EM structure of one chimeric VLP (Yokote/Mc114) was solved to 8.5-Å resolution. A homology model of the VP1 indicated for the first time the putative S and P (P1-1, P2, and P1-2) domains. The overall structure of Yokote/Mc114 contained features common among other caliciviruses. We showed that the P2 subdomain was mainly involved in the homodimeric interface, whereas a large gap between the P1 subdomains had fewer interactions.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures









References
-
- Hansman GS, Jiang XJ, Green KY. 2010. Caliciviruses: molecular and cellular virology. Caister Academic Press, Norfolk, United Kingdom.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

