Culture-Based and Culture-Independent Bacteriologic Analysis of Cystic Fibrosis Respiratory Specimens
- PMID: 26699705
- PMCID: PMC4767965
- DOI: 10.1128/JCM.02299-15
Culture-Based and Culture-Independent Bacteriologic Analysis of Cystic Fibrosis Respiratory Specimens
Abstract
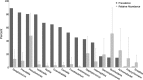
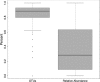
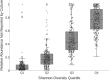
Cystic fibrosis (CF) is characterized by chronic infection and inflammation of the airways. In vitro culture of select bacterial species from respiratory specimens has been used to guide antimicrobial therapy in CF for the past few decades. More recently, DNA sequence-based, culture-independent approaches have been used to assess CF airway microbiology, although the role that these methods will (or should) have in routine microbiologic analysis of CF respiratory specimens is unclear. We performed DNA sequence analyses to detect bacterial species in 945 CF sputum samples that had been previously analyzed by selective CF culture. We determined the concordance of results based on culture and sequence analysis, highlighting the comparison of the results for the most prevalent genera. Although overall prevalence rates were comparable between the two methods, results varied by genus. While sequence analysis was more likely to detect Achromobacter, Stenotrophomonas, and Burkholderia, it was less likely to detect Staphylococcus. Streptococcus spp. were rarely reported in culture results but were the most frequently detected species by sequence analysis. A variety of obligate and facultative anaerobic species, not reported by culture, was also detected with high prevalence by sequence analysis. Sequence analysis indicated that in a considerable proportion of samples, taxa not reported by selective culture constituted a relatively high proportion of the total bacterial load, suggesting that routine CF culture may underrepresent significant segments of the bacterial communities inhabiting CF airways.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Hansen SK, Rau MH, Johansen HK, Ciofu O, Jelsbak L, Yang L, Folkesson A, Jarmer HO, Aanaes K, von Buchwald C, Hoiby N, Molin S. 2012. Evolution and diversification of Pseudomonas aeruginosa in the paranasal sinuses of cystic fibrosis children have implications for chronic lung infection. ISME J 6:31–45. doi: 10.1038/ismej.2011.83. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

