Evolutionary loss of peroxisomes--not limited to parasites
- PMID: 26700421
- PMCID: PMC4690255
- DOI: 10.1186/s13062-015-0101-6
Evolutionary loss of peroxisomes--not limited to parasites
Abstract
Background: Peroxisomes are ubiquitous eukaryotic organelles that compartmentalize a variety of metabolic pathways that are primarily related to the oxidative metabolism of lipids and the detoxification of reactive oxygen species. The importance of peroxisomes is underscored by serious human diseases, which are caused by disorders in peroxisomal functions. Some eukaryotic lineages, however, lost peroxisomes. These organisms are mainly anaerobic protists and some parasitic lineages including Plasmodium and parasitic platyhelminths. Here we performed a systematic in-silico analysis of peroxisomal markers among metazoans to assess presence of peroxisomes and peroxisomal enzymes.
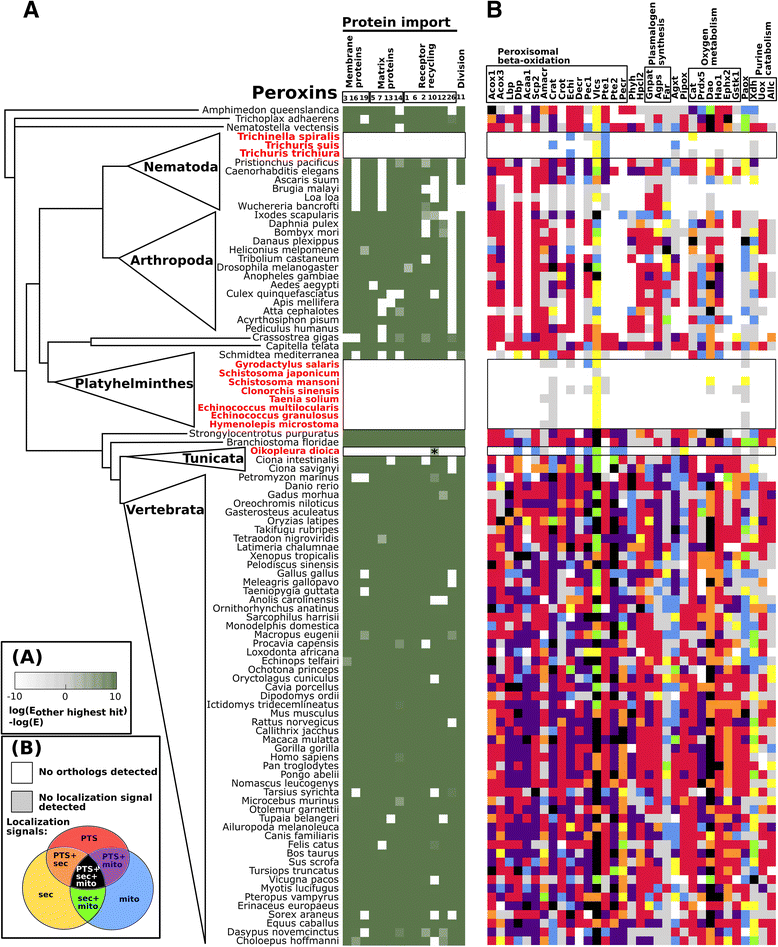
Results: Our analyses reveal an obvious loss of peroxisomes in all tested flukes, tapeworms, and parasitic roundworms of the order Trichocephalida. Intriguingly, peroxisomal markers are absent from the genome of the free-living tunicate Oikopleura dioica, which inhabits oxygen-containing niches of sea waters. We further map the presence and predicted subcellular localization of putative peroxisomal enzymes, showing that in organisms without the peroxisomal markers the set of these enzymes is highly reduced and none of them contains a predicted peroxisomal targeting signal.
Conclusions: We have shown that several lineages of metazoans independently lost peroxisomes and that the loss of peroxisomes was not exclusively associated with adaptation to anaerobic habitats and a parasitic lifestyle. Although the reason for the loss of peroxisomes from O. dioica is unclear, organisms lacking peroxisomes, including the free-living O. dioica, share certain typical r-selected traits: high fecundity, limited ontogenesis and relatively low complexity of the gene content. We hypothesize that peroxisomes are generally the first compartment to be lost during evolutionary reductions of the eukaryotic cell.
Figures

Similar articles
-
The evolutionary origin of peroxisomes: an ER-peroxisome connection.Mol Biol Evol. 2006 Apr;23(4):838-45. doi: 10.1093/molbev/msj103. Epub 2006 Feb 1. Mol Biol Evol. 2006. PMID: 16452116
-
Peroxisomes in parasitic protists.Mol Biochem Parasitol. 2016 Sep-Oct;209(1-2):35-45. doi: 10.1016/j.molbiopara.2016.02.005. Epub 2016 Feb 16. Mol Biochem Parasitol. 2016. PMID: 26896770 Review.
-
Distribution and Evolution of Peroxisomes in Alveolates (Apicomplexa, Dinoflagellates, Ciliates).Genome Biol Evol. 2018 Jan 1;10(1):1-13. doi: 10.1093/gbe/evx250. Genome Biol Evol. 2018. PMID: 29202176 Free PMC article.
-
[Peroxisomal diseases--oxygen and free radicals].Wien Klin Wochenschr. 1995;107(22):690-3. Wien Klin Wochenschr. 1995. PMID: 8533430 Review. German.
-
The evolution of eukaryotic cells from the perspective of peroxisomes: phylogenetic analyses of peroxisomal beta-oxidation enzymes support mitochondria-first models of eukaryotic cell evolution.Bioessays. 2015 Feb;37(2):195-203. doi: 10.1002/bies.201400151. Epub 2014 Nov 13. Bioessays. 2015. PMID: 25394329
Cited by
-
Comparative Genomics of Peroxisome Biogenesis Proteins: Making Sense of the PEX Proteins.Front Cell Dev Biol. 2021 May 20;9:654163. doi: 10.3389/fcell.2021.654163. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34095119 Free PMC article.
-
Phylogenomic Analysis of 155 Helminth Species Reveals Widespread Absence of Oxygen Metabolic Capacity.Genome Biol Evol. 2023 Aug 1;15(8):evad135. doi: 10.1093/gbe/evad135. Genome Biol Evol. 2023. PMID: 37481257 Free PMC article.
-
Anaerobic peroxisomes in Mastigamoeba balamuthi.Proc Natl Acad Sci U S A. 2020 Jan 28;117(4):2065-2075. doi: 10.1073/pnas.1909755117. Epub 2020 Jan 13. Proc Natl Acad Sci U S A. 2020. PMID: 31932444 Free PMC article.
-
Functional Analyses of a Putative, Membrane-Bound, Peroxisomal Protein Import Mechanism from the Apicomplexan Protozoan Toxoplasma gondii.Genes (Basel). 2018 Aug 29;9(9):434. doi: 10.3390/genes9090434. Genes (Basel). 2018. PMID: 30158461 Free PMC article.
-
Telomere-to-telomere assembly of the genome of an individual Oikopleura dioica from Okinawa using Nanopore-based sequencing.BMC Genomics. 2021 Mar 29;22(1):222. doi: 10.1186/s12864-021-07512-6. BMC Genomics. 2021. PMID: 33781200 Free PMC article.
References
-
- Rucktäschel R, Girzalsky W, Erdmann R. Protein import machineries of peroxisomes. Biochim Biophys Acta BBA - Biomembr. 1808;2011:892–900. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

