The function and regulation of the GATA factor ELT-2 in the C. elegans endoderm
- PMID: 26700680
- PMCID: PMC4760314
- DOI: 10.1242/dev.130914
The function and regulation of the GATA factor ELT-2 in the C. elegans endoderm
Abstract
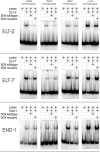
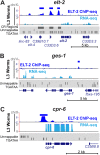
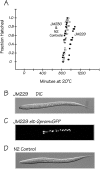
ELT-2 is the major regulator of genes involved in differentiation, maintenance and function of C. elegans intestine from the early embryo to mature adult. elt-2 responds to overexpression of the GATA transcription factors END-1 and END-3, which specify the intestine, as well as to overexpression of the two GATA factors that are normally involved in intestinal differentiation, ELT-7 and ELT-2 itself. Little is known about the molecular mechanisms underlying these interactions, how ELT-2 levels are maintained throughout development or how such systems respond to developmental perturbations. Here, we analyse elt-2 gene regulation through transgenic reporter assays, ELT-2 ChIP and characterisation of in vitro DNA-protein interactions. Our results indicate that elt-2 is controlled by three discrete regulatory regions conserved between C. elegans and C. briggsae that span >4 kb of 5' flanking sequence. These regions are superficially interchangeable but have quantitatively different enhancer properties, and their combined activities indicate inter-region synergies. Their regulatory activity is mediated by a small number of conserved TGATAA sites that are largely interchangeable and interact with different endodermal GATA factors with only modest differences in affinity. The redundant molecular mechanism that forms the elt-2 regulatory network is robust and flexible, as loss of end-3 halves ELT-2 levels in the early embryo but levels fully recover by the time of hatching. When ELT-2 is expressed under the control of end-1 regulatory elements, in addition to its own endogenous promoter, it can replace the complete set of endoderm-specific GATA factors: END-1, END-3, ELT-7 and (the probably non-functional) ELT-4. Thus, in addition to controlling gene expression during differentiation, ELT-2 is capable of specifying the entire C. elegans endoderm.
Keywords: Caenorhabditis elegans; ChIP-Seq; ELT-2; Endoderm development; GATA factor; Transcription.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


Comment in
-
Probing and rearranging the transcription factor network controlling the C. elegans endoderm.Worm. 2016 Jun 10;5(3):e1198869. doi: 10.1080/21624054.2016.1198869. eCollection 2016. Worm. 2016. PMID: 27695655 Free PMC article.
Similar articles
-
A Strategy To Isolate Modifiers of Caenorhabditis elegans Lethal Mutations: Investigating the Endoderm Specifying Ability of the Intestinal Differentiation GATA Factor ELT-2.G3 (Bethesda). 2018 May 4;8(5):1425-1437. doi: 10.1534/g3.118.200079. G3 (Bethesda). 2018. PMID: 29593072 Free PMC article.
-
Quantitating transcription factor redundancy: The relative roles of the ELT-2 and ELT-7 GATA factors in the C. elegans endoderm.Dev Biol. 2018 Mar 15;435(2):150-161. doi: 10.1016/j.ydbio.2017.12.023. Epub 2018 Jan 31. Dev Biol. 2018. PMID: 29360433 Free PMC article.
-
Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and -7 mediates the specification→differentiation transition.Dev Biol. 2010 Nov 1;347(1):154-66. doi: 10.1016/j.ydbio.2010.08.020. Epub 2010 Aug 31. Dev Biol. 2010. PMID: 20807527 Free PMC article.
-
The Caenorhabditis elegans intestine.Wiley Interdiscip Rev Dev Biol. 2013 May-Jun;2(3):347-67. doi: 10.1002/wdev.93. Epub 2012 Oct 9. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 23799580 Review.
-
GATA factors as key regulatory molecules in the development of Drosophila endoderm.Dev Growth Differ. 2005 Dec;47(9):581-9. doi: 10.1111/j.1440-169X.2005.00836.x. Dev Growth Differ. 2005. PMID: 16316403 Review.
Cited by
-
Feedforward regulatory logic controls the specification-to-differentiation transition and terminal cell fate during Caenorhabditis elegans endoderm development.Development. 2022 Jun 15;149(12):dev200337. doi: 10.1242/dev.200337. Epub 2022 Jun 27. Development. 2022. PMID: 35758255 Free PMC article.
-
The long isoform of the C. elegans ELT-3 GATA factor can specify endoderm when overexpressed.MicroPubl Biol. 2023 Jan 21;2023:10.17912/micropub.biology.000748. doi: 10.17912/micropub.biology.000748. eCollection 2023. MicroPubl Biol. 2023. PMID: 36748041 Free PMC article.
-
Evolutionary Dynamics of the SKN-1 → MED → END-1,3 Regulatory Gene Cascade in Caenorhabditis Endoderm Specification.G3 (Bethesda). 2020 Jan 7;10(1):333-356. doi: 10.1534/g3.119.400724. G3 (Bethesda). 2020. PMID: 31740453 Free PMC article.
-
The C. elegans GATA transcription factor elt-2 mediates distinct transcriptional responses and opposite infection outcomes towards different Bacillus thuringiensis strains.PLoS Pathog. 2020 Sep 24;16(9):e1008826. doi: 10.1371/journal.ppat.1008826. eCollection 2020 Sep. PLoS Pathog. 2020. PMID: 32970778 Free PMC article.
-
Probing and rearranging the transcription factor network controlling the C. elegans endoderm.Worm. 2016 Jun 10;5(3):e1198869. doi: 10.1080/21624054.2016.1198869. eCollection 2016. Worm. 2016. PMID: 27695655 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

