DJ Pairing during VDJ Recombination Shows Positional Biases That Vary among Individuals with Differing IGHD Locus Immunogenotypes
- PMID: 26700767
- PMCID: PMC4724508
- DOI: 10.4049/jimmunol.1501401
DJ Pairing during VDJ Recombination Shows Positional Biases That Vary among Individuals with Differing IGHD Locus Immunogenotypes
Abstract
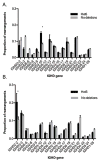
Human IgH diversity is influenced by biases in the pairing of IGHD and IGHJ genes, but these biases have not been described in detail. We used high-throughput sequencing of VDJ rearrangements to explore DJ pairing biases in 29 individuals. It was possible to infer three contrasting IGHD-IGHJ haplotypes in nine of these individuals, and two of these haplotypes include deletion polymorphisms involving multiple contiguous IGHD genes. Therefore, we were able to explore how the underlying genetic makeup of the H chain locus influences the formation of particular DJ pairs. Analysis of nonproductive rearrangements demonstrates that 3' IGHD genes tend to pair preferentially with 5' IGHJ genes, whereas 5' IGHD genes pair preferentially with 3' IGHJ genes; the relationship between IGHD gene pairing frequencies and IGHD gene position is a near linear one for each IGHJ gene. However, striking differences are seen in individuals who carry deletion polymorphisms in the D locus. The absence of different blocks of IGHD genes leads to increases in the utilization frequencies of just a handful of genes, and these genes have no clear positional relationships to the deleted genes. This suggests that pairing frequencies may be influenced by additional complex positional relationships that perhaps arise from chromatin structure. In contrast to IGHD gene usage, IGHJ gene usage is unaffected by the IGHD gene-deletion polymorphisms. Such an outcome would be expected if the recombinase complex associates with an IGHJ gene before associating with an IGHD gene partner.
Copyright © 2016 by The American Association of Immunologists, Inc.
Figures
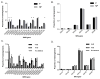

 IGHJ4 ▲IGHJ5 X IGHJ6
IGHJ4 ▲IGHJ5 X IGHJ6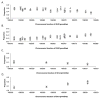
References
-
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. - PubMed
-
- Matsuda F, Shin EK, Nagaoka H, Matsumura R, Haino M, Fukita Y, Taka-ishi S, Imai T, Riley JH, Anand R, et al. Structure and physical map of 64 variable segments in the 3’0.8-megabase region of the human immunoglobulin heavy-chain locus. Nat Genet. 1993;3:88–94. - PubMed
-
- Boyd SD, Gaeta BA, Jackson KJ, Fire AZ, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Collins AM. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol. 2010;184:6986–6992. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

