Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss
- PMID: 26701265
- PMCID: PMC4701977
- DOI: 10.1101/gad.270959.115
Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss
Abstract
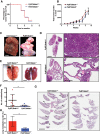
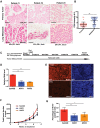
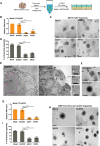
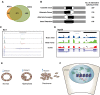
Genome-wide analyses have identified thousands of long noncoding RNAs (lncRNAs). Malat1 (metastasis-associated lung adenocarcinoma transcript 1) is among the most abundant lncRNAs whose expression is altered in numerous cancers. Here we report that genetic loss or systemic knockdown of Malat1 using antisense oligonucleotides (ASOs) in the MMTV (mouse mammary tumor virus)-PyMT mouse mammary carcinoma model results in slower tumor growth accompanied by significant differentiation into cystic tumors and a reduction in metastasis. Furthermore, Malat1 loss results in a reduction of branching morphogenesis in MMTV-PyMT- and Her2/neu-amplified tumor organoids, increased cell adhesion, and loss of migration. At the molecular level, Malat1 knockdown results in alterations in gene expression and changes in splicing patterns of genes involved in differentiation and protumorigenic signaling pathways. Together, these data demonstrate for the first time a functional role of Malat1 in regulating critical processes in mammary cancer pathogenesis. Thus, Malat1 represents an exciting therapeutic target, and Malat1 ASOs represent a potential therapy for inhibiting breast cancer progression.
Keywords: Malat1; antisense therapy; breast cancer; metastasis; noncoding RNA; tumor differentiation; tumor organoids.
© 2016 Arun et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Ades F, Zardavas D, Bozovic-Spasojevic I, Pugliano L, Fumagalli D, de Azambuja E, Viale G, Sotiriou C, Piccart M. 2014. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J Clin Oncol 32: 2794–2803. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
