TrpE feedback mutants reveal roadblocks and conduits toward increasing secondary metabolism in Aspergillus fumigatus
- PMID: 26701311
- PMCID: PMC4789178
- DOI: 10.1016/j.fgb.2015.12.002
TrpE feedback mutants reveal roadblocks and conduits toward increasing secondary metabolism in Aspergillus fumigatus
Abstract
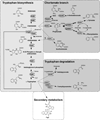
Small peptides formed from non-ribosomal peptide synthetases (NRPS) are bioactive molecules produced by many fungi including the genus Aspergillus. A subset of NRPS utilizes tryptophan and its precursor, the non-proteinogenic amino acid anthranilate, in synthesis of various metabolites such as Aspergillus fumigatus fumiquinazolines (Fqs) produced by the fmq gene cluster. The A. fumigatus genome contains two putative anthranilate synthases - a key enzyme in conversion of anthranilic acid to tryptophan - one beside the fmq cluster and one in a region of co-linearity with other Aspergillus spp. Only the gene found in the co-linear region, trpE, was involved in tryptophan biosynthesis. We found that site-specific mutations of the TrpE feedback domain resulted in significantly increased production of anthranilate, tryptophan, p-aminobenzoate and fumiquinazolines FqF and FqC. Supplementation with tryptophan restored metabolism to near wild type levels in the feedback mutants and suggested that synthesis of the tryptophan degradation product kynurenine could negatively impact Fq synthesis. The second putative anthranilate synthase gene next to the fmq cluster was termed icsA for its considerable identity to isochorismate synthases in bacteria. Although icsA had no impact on A. fumigatus Fq production, deletion and over-expression of icsA increased and decreased respectively aromatic amino acid levels suggesting that IcsA can draw from the cellular chorismate pool.
Keywords: Aspergillus fumigatus; Fumiquinazoline; Metabolic engineering; Metabolic flux; Secondary metabolism; Tryptophan metabolism.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



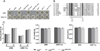
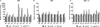

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

