The significance of Brf1 overexpression in human hepatocellular carcinoma
- PMID: 26701855
- PMCID: PMC4868753
- DOI: 10.18632/oncotarget.6668
The significance of Brf1 overexpression in human hepatocellular carcinoma
Abstract
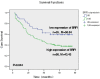
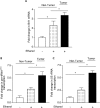
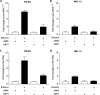
Brf1 (TFIIB-related factor 1) plays a crucial role in cell transformation and tumorigenesis. However, the significance of Brf1 expression in human HCC (hepatocellular carcinoma) cases remains to be addressed. In this study, biopsies of human HCC, liver tumor samples of mice and cell lines of normal and tumor liver were utilized to determine the alteration of Brf1 expression using cytological and molecular biological approaches. Brf1 expression is increased in human HCC cases, which is correlated with shorter survival times. Levels of Brf1 and Pol III (RNA polymerase III-dependent) gene transcription in HCC patients with alcohol consumption are higher than the cases of non-HCC with or without alcohol intake. Induction of Brf1 and Pol III genes by ethanol in hepatoma cells is higher than in non-tumor cells. Ethanol increases the rate of cell transformation. Repression of Brf1 inhibits alcohol-promoted cell transformation. Alcohol consumption enhances Brf1 expression to promote cell transformation. These studies demonstrate that Brf1 is a new biomarker of HCC.
Keywords: Brf1; Pol III genes; alcohol; hepatocellular carcinoma; survival.
Conflict of interest statement
The authors who have taken part in this study declared that they do not have anything to disclose regarding funding or conflict of interest with respect to this manuscript.
Figures




References
-
- Lieber C. Hepatic, metabolic, and nutritional disorders of alcoholism: from pathogenesis to therapy. Crit Rev Clin Lab Sci. 2000;37:551–584. - PubMed
-
- Seitz HK, Poschl G, Simanowski UA. Alcohol and cancer. Recent Dev Alcohol. 1998;14:67–95. - PubMed
-
- Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer. 2007;7:599–612. - PubMed
-
- Yuan JM, Govindarajan S, Arakawa K, Yu MC. Synergism of alcohol, diabetes, and viral hepatitis on the risk of hepatocellular carcinoma in blacks and whites in the U.S. Cancer. 2004;101:1009–1017. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

