Targetable genetic features of primary testicular and primary central nervous system lymphomas
- PMID: 26702065
- PMCID: PMC4760091
- DOI: 10.1182/blood-2015-10-673236
Targetable genetic features of primary testicular and primary central nervous system lymphomas
Abstract
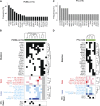
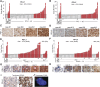
Primary central nervous system lymphomas (PCNSLs) and primary testicular lymphomas (PTLs) are extranodal large B-cell lymphomas (LBCLs) with inferior responses to current empiric treatment regimens. To identify targetable genetic features of PCNSL and PTL, we characterized their recurrent somatic mutations, chromosomal rearrangements, copy number alterations (CNAs), and associated driver genes, and compared these comprehensive genetic signatures to those of diffuse LBCL and primary mediastinal large B-cell lymphoma (PMBL). These studies identify unique combinations of genetic alterations in discrete LBCL subtypes and subtype-selective bases for targeted therapy. PCNSLs and PTLs frequently exhibit genomic instability, and near-uniform, often biallelic, CDKN2A loss with rare TP53 mutations. PCNSLs and PTLs also use multiple genetic mechanisms to target key genes and pathways and exhibit near-uniform oncogenic Toll-like receptor signaling as a result of MYD88 mutation and/or NFKBIZ amplification, frequent concurrent B-cell receptor pathway activation, and deregulation of BCL6. Of great interest, PCNSLs and PTLs also have frequent 9p24.1/PD-L1/PD-L2 CNAs and additional translocations of these loci, structural bases of immune evasion that are shared with PMBL.
© 2016 by The American Society of Hematology.
Figures





References
-
- Cheah CY, Wirth A, Seymour JF. Primary testicular lymphoma. Blood. 2014;123(4):486–493. - PubMed
-
- Horne MJ, Adeniran AJ. Primary diffuse large B-cell lymphoma of the testis. Arch Pathol Lab Med. 2011;135(10):1363–1367. - PubMed
-
- Deckert M, Montesinos-Rongen M, Brunn A, Siebert R. Systems biology of primary CNS lymphoma: from genetic aberrations to modeling in mice. Acta Neuropathol. 2014;127(2):175–188. - PubMed
-
- Riemersma SA, Jordanova ES, Schop RF, et al. Extensive genetic alterations of the HLA region, including homozygous deletions of HLA class II genes in B-cell lymphomas arising in immune-privileged sites. Blood. 2000;96(10):3569–3577. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

