UCH-L1 is induced in germinal center B cells and identifies patients with aggressive germinal center diffuse large B-cell lymphoma
- PMID: 26702068
- PMCID: PMC4807423
- DOI: 10.1182/blood-2015-07-656678
UCH-L1 is induced in germinal center B cells and identifies patients with aggressive germinal center diffuse large B-cell lymphoma
Abstract
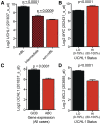
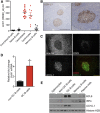
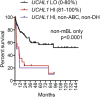
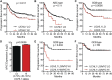
Gene expression profiling has identified 2 major subclasses of diffuse large B-cell lymphoma (DLBCL). Cases resembling germinal center (GC) B cells (GCB-DLBCL) generally occur in younger patients, have a distinct molecular pathophysiology, and have improved outcomes compared with those similar to activated post-GC cells (activated B-cell DLBCL). We previously found that the ubiquitin hydrolase UCH-L1 is frequently overexpressed in mature B-cell malignancies and is a potent oncogene in mice. The cause for its overexpression in lymphoma, and whether it impacts the outcome of patients with DLBCL is unknown. Here, we show that UCH-L1 reflects GC lineage in lymphoma and is an oncogenic biomarker of aggressive GCB-DLBCL. We find that UCH-L1 is specifically induced in GC B cells in mice and humans, and that its expression correlates highly with the GCB subtype in DLBCL. We also find that UCH-L1 cooperates with BCL6 in a mouse model of GC B-cell lymphoma, but not with the development of multiple myeloma derived from post-GC cells. Despite the typically good outcomes of GCB-DLBCL, increased UCHL1 identifies a subgroup with early relapses independent of MYC expression, suggesting biological diversity in this subset of disease. Consistent with this, forced Uchl1 overexpression had a substantial impact on gene expression in GC B cells including pathways of cell cycle progression, cell death and proliferation, and DNA replication. These data demonstrate a novel role for UCH-L1 outside of the nervous system and suggest its potential use as a biomarker and therapeutic target in DLBCL.
© 2016 by The American Society of Hematology.
Figures


Comment in
-
UCH-L1 in DLBCL: marker or target?Blood. 2016 Mar 24;127(12):1524-5. doi: 10.1182/blood-2016-01-689984. Blood. 2016. PMID: 27013211 Free PMC article.
Similar articles
-
UCH-L1 in DLBCL: marker or target?Blood. 2016 Mar 24;127(12):1524-5. doi: 10.1182/blood-2016-01-689984. Blood. 2016. PMID: 27013211 Free PMC article.
-
Loss of signalling via Gα13 in germinal centre B-cell-derived lymphoma.Nature. 2014 Dec 11;516(7530):254-8. doi: 10.1038/nature13765. Epub 2014 Sep 28. Nature. 2014. PMID: 25274307 Free PMC article.
-
MYC/BCL6 double-hit lymphoma (DHL): a tumour associated with an aggressive clinical course and poor prognosis.Histopathology. 2016 Jun;68(7):1090-8. doi: 10.1111/his.12884. Epub 2015 Dec 8. Histopathology. 2016. PMID: 26426741
-
Molecular genetics of aggressive B-cell lymphoma.Hematol Oncol. 2017 Jun;35 Suppl 1:76-79. doi: 10.1002/hon.2405. Hematol Oncol. 2017. PMID: 28591417 Review. No abstract available.
-
Diffuse large B-cell lymphoma.Pathology. 2018 Jan;50(1):74-87. doi: 10.1016/j.pathol.2017.09.006. Epub 2017 Nov 20. Pathology. 2018. PMID: 29167021 Review.
Cited by
-
DUBbing Down Translation: The Functional Interaction of Deubiquitinases with the Translational Machinery.Mol Cancer Ther. 2019 Sep;18(9):1475-1483. doi: 10.1158/1535-7163.MCT-19-0307. Mol Cancer Ther. 2019. PMID: 31481479 Free PMC article. Review.
-
Diffuse large B-cell lymphoma: R-CHOP failure-what to do?Hematology Am Soc Hematol Educ Program. 2016 Dec 2;2016(1):366-378. doi: 10.1182/asheducation-2016.1.366. Hematology Am Soc Hematol Educ Program. 2016. PMID: 27913503 Free PMC article. Review.
-
Ubiquitin Carboxyl-Terminal Hydrolases and Human Malignancies: The Novel Prognostic and Therapeutic Implications for Head and Neck Cancer.Front Oncol. 2021 Jan 29;10:592501. doi: 10.3389/fonc.2020.592501. eCollection 2020. Front Oncol. 2021. PMID: 33585209 Free PMC article. Review.
-
Establishment of an ovarian cancer omentum metastasis-related prognostic model by integrated analysis of scRNA-seq and bulk RNA-seq.J Ovarian Res. 2022 Nov 23;15(1):123. doi: 10.1186/s13048-022-01059-0. J Ovarian Res. 2022. PMID: 36424614 Free PMC article.
-
The deubiquitinating enzyme UCHL1 is a favorable prognostic marker in neuroblastoma as it promotes neuronal differentiation.J Exp Clin Cancer Res. 2018 Oct 25;37(1):258. doi: 10.1186/s13046-018-0931-z. J Exp Clin Cancer Res. 2018. PMID: 30359286 Free PMC article.
References
-
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. - PubMed
-
- Friedberg JW. Relapsed/refractory diffuse large B-cell lymphoma. Hematology Am Soc Hematol Educ Program. 2011;2011:498–505. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

