The E3 Ubiquitin Ligase TRIM9 Is a Filopodia Off Switch Required for Netrin-Dependent Axon Guidance
- PMID: 26702829
- PMCID: PMC4707677
- DOI: 10.1016/j.devcel.2015.11.022
The E3 Ubiquitin Ligase TRIM9 Is a Filopodia Off Switch Required for Netrin-Dependent Axon Guidance
Abstract
Neuronal growth cone filopodia contain guidance receptors and contribute to axon guidance; however, the mechanism by which the guidance cue netrin increases filopodia density is unknown. Here, we demonstrate that TRIM9, an E3 ubiquitin ligase that localizes to filopodia tips and binds the netrin receptor DCC, interacts with and ubiquitinates the barbed-end polymerase VASP to modulate filopodial stability during netrin-dependent axon guidance. Studies with murine Trim9(+/+) and Trim9(-/-) cortical neurons, along with a non-ubiquitinatable VASP mutant, demonstrate that TRIM9-mediated ubiquitination of VASP reduces VASP filopodial tip localization, VASP dynamics at tips, and filopodial stability. Upon netrin treatment, VASP is deubiquitinated, which promotes VASP tip localization and filopodial stability. Trim9 deletion induces axon guidance defects in vitro and in vivo, whereas a gradient of deubiquitinase inhibition promotes axon turning in vitro. We conclude that a gradient of TRIM9-mediated ubiquitination of VASP creates a filopodial stability gradient during axon turning.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
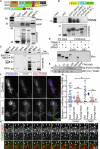

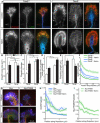
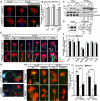
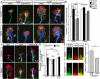

Comment in
-
New Light on Growth Cone Navigation.Dev Cell. 2015 Dec 21;35(6):672-3. doi: 10.1016/j.devcel.2015.12.002. Dev Cell. 2015. PMID: 26702827
References
-
- Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ, Kumar KGS, Konietzny R, Fischer R, Kogan E, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 2011;18:1401–1412. - PubMed
-
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM108970/GM/NIGMS NIH HHS/United States
- R01 GM068678/GM/NIGMS NIH HHS/United States
- F31 NS087837-01A1/NS/NINDS NIH HHS/United States
- F31 NS087837/NS/NINDS NIH HHS/United States
- T32 NS007431/NS/NINDS NIH HHS/United States
- K12HD073945/HD/NICHD NIH HHS/United States
- R42 MH097377/MH/NIMH NIH HHS/United States
- R41MH097377/MH/NIMH NIH HHS/United States
- K12 HD073945/HD/NICHD NIH HHS/United States
- R41 MH097377/MH/NIMH NIH HHS/United States
- GM108970/GM/NIGMS NIH HHS/United States
- T32 NS007431-16/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
