A multistate tuberculosis pharmacometric model: a framework for studying anti-tubercular drug effects in vitro
- PMID: 26702921
- PMCID: PMC4790616
- DOI: 10.1093/jac/dkv416
A multistate tuberculosis pharmacometric model: a framework for studying anti-tubercular drug effects in vitro
Abstract
Objectives: Mycobacterium tuberculosis can exist in different states in vitro, which can be denoted as fast multiplying, slow multiplying and non-multiplying. Characterizing the natural growth of M. tuberculosis could provide a framework for accurate characterization of drug effects on the different bacterial states.
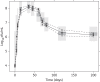
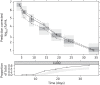
Methods: The natural growth data of M. tuberculosis H37Rv used in this study consisted of viability defined as cfu versus time based on data from an in vitro hypoxia system. External validation of the natural growth model was conducted using data representing the rate of incorporation of radiolabelled methionine into proteins by the bacteria. Rifampicin time-kill curves from log-phase (0.25-16 mg/L) and stationary-phase (0.5-64 mg/L) cultures were used to assess the model's ability to describe drug effects by evaluating different linear and non-linear exposure-response relationships.
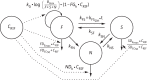
Results: The final pharmacometric model consisted of a three-compartment differential equation system representing fast-, slow- and non-multiplying bacteria. Model predictions correlated well with the external data (R(2) = 0.98). The rifampicin effects on log-phase and stationary-phase cultures were separately and simultaneously described by including the drug effect on the different bacterial states. The predicted reduction in log10 cfu after 14 days and at 0.5 mg/L was 2.2 and 0.8 in the log-phase and stationary-phase systems, respectively.
Conclusions: The model provides predictions of the change in bacterial numbers for the different bacterial states with and without drug effect and could thus be used as a framework for studying anti-tubercular drug effects in vitro.
© The Author 2015. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy.
Figures




References
-
- World Health Organization. Global Tuberculosis Report 2014. http://apps.who.int/iris/bitstream/10665/137094/1/9789241564809_eng.pdf.
-
- Okusanya OO, Tsuji BT, Bülitta JB et al. . Evaluation of the pharmacokinetics-pharmacodynamics of fusidic acid against Staphylococcus aureus and Streptococcus pyogenes using in vitro infection models: implications for dose selection. Diagn Microbiol Infect Dis 2011; 70: 101–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

