Genome-wide analysis of WRKY transcription factors in white pear (Pyrus bretschneideri) reveals evolution and patterns under drought stress
- PMID: 26704366
- PMCID: PMC4691019
- DOI: 10.1186/s12864-015-2233-6
Genome-wide analysis of WRKY transcription factors in white pear (Pyrus bretschneideri) reveals evolution and patterns under drought stress
Abstract
Background: WRKY transcription factors (TFs) constitute one of the largest protein families in higher plants, and its members contain one or two conserved WRKY domains, about 60 amino acid residues with the WRKYGQK sequence followed by a C2H2 or C2HC zinc finger motif. WRKY proteins play significant roles in plant development, and in responses to biotic and abiotic stresses. Pear (Pyrus bretschneideri) is one of the most important fruit crops in the world and is frequently threatened by abiotic stress, such as drought, affecting growth, development and productivity. Although the pear genome sequence has been released, little is known about the WRKY TFs in pear, especially in respond to drought stress at the genome-wide level.
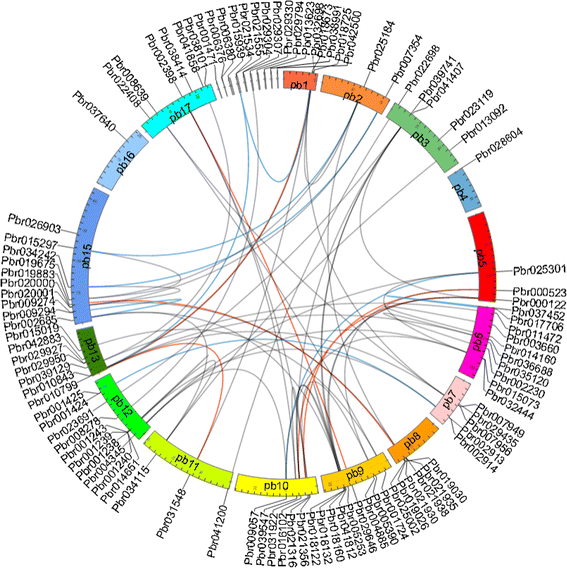
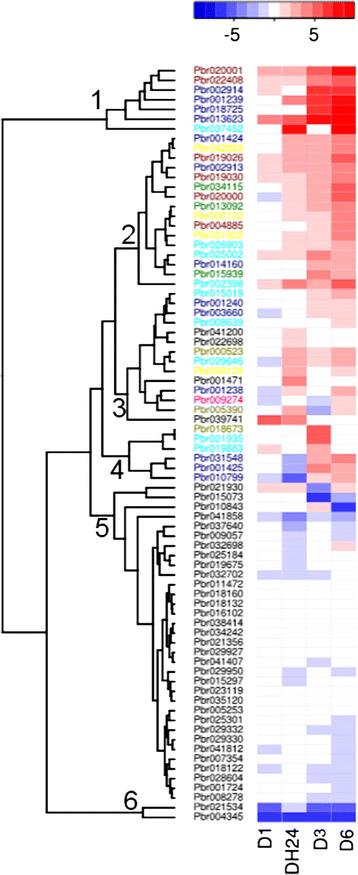
Results: We identified a total of 103 WRKY TFs in the pear genome. Based on the structural features of WRKY proteins and topology of the phylogenetic tree, the pear WRKY (PbWRKY) family was classified into seven groups (Groups 1, 2a-e, and 3). The microsyteny analysis indicated that 33 (32%) PbWRKY genes were tandemly duplicated and 57 genes (55.3%) were segmentally duplicated. RNA-seq experiment data and quantitative real-time reverse transcription PCR revealed that PbWRKY genes in different groups were induced by drought stress, and Group 2a and 3 were mainly involved in the biological pathways in response to drought stress. Furthermore, adaptive evolution analysis detected a significant positive selection for Pbr001425 in Group 3, and its expression pattern differed from that of other members in this group. The present study provides a solid foundation for further functional dissection and molecular evolution of WRKY TFs in pear, especially for improving the water-deficient resistance of pear through manipulation of the PbWRKYs.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

