MycoBASE: expanding the functional annotation coverage of mycobacterial genomes
- PMID: 26704706
- PMCID: PMC4690229
- DOI: 10.1186/s12864-015-2311-9
MycoBASE: expanding the functional annotation coverage of mycobacterial genomes
Abstract
Background: Central to most omic scale experiments is the interpretation and examination of resulting gene lists corresponding to differentially expressed, regulated, or observed gene or protein sets. Complicating interpretation is a lack of functional annotation assigned to a large percentage of many microbial genomes. This is particularly noticeable in mycobacterial genomes, which are significantly divergent from many of the microbial model species used for gene and protein functional characterization, but which are extremely important clinically. Mycobacterial species, ranging from M. tuberculosis to M. abscessus, are responsible for deadly infectious diseases that kill over 1.5 million people each year across the world. A better understanding of the coding capacity of mycobacterial genomes is therefore necessary to shed increasing light on putative mechanisms of virulence, pathogenesis, and functional adaptations.
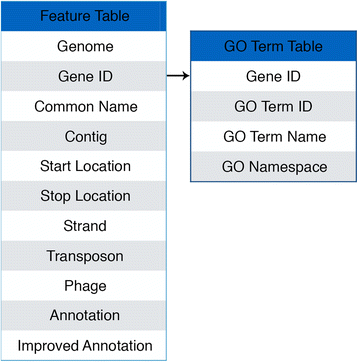
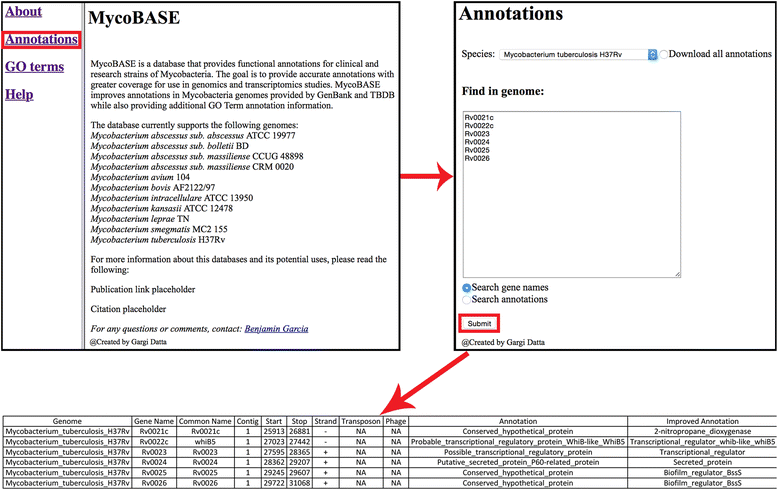
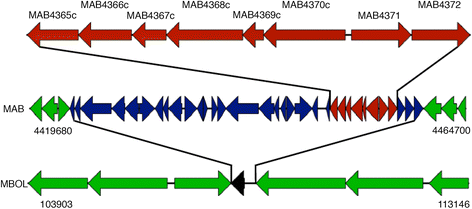
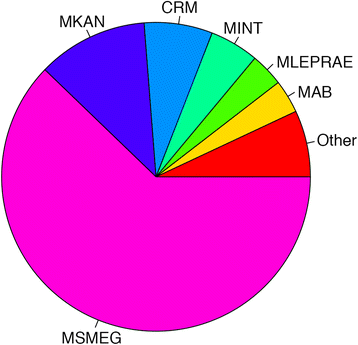
Description: Here we describe the improved functional annotation coverage of 11 important mycobacterial genomes, many involved in human diseases including tuberculosis, leprosy, and nontuberculous mycobacterial (NTM) infections. Of the 11 mycobacterial genomes, we provide 9899 new functional annotations, compared to NCBI and TBDB annotations, for genes previously characterized as genes of unknown function, hypothetical, and hypothetical conserved proteins. Functional annotations are available at our newly developed web resource MycoBASE (Mycobacterial Annotation Server) at strong.ucdenver.edu/mycobase.
Conclusion: Improved annotations allow for better understanding and interpretation of genomic and transcriptomic experiments, including analyzing the functional implications of insertions, deletions, and mutations, inferring the function of understudied genes, and determining functional changes resulting from differential expression studies. MycoBASE provides a valuable resource for mycobacterial researchers, through improved and searchable functional annotations and functional enrichment strategies. MycoBASE will be continually supported and updated to include new genomes, enabling a powerful resource to aid the quest to better understand these important pathogenic and environmental species.
Figures



Similar articles
-
MycoCAP - Mycobacterium Comparative Analysis Platform.Sci Rep. 2015 Dec 15;5:18227. doi: 10.1038/srep18227. Sci Rep. 2015. PMID: 26666970 Free PMC article.
-
proGenomes: a resource for consistent functional and taxonomic annotations of prokaryotic genomes.Nucleic Acids Res. 2017 Jan 4;45(D1):D529-D534. doi: 10.1093/nar/gkw989. Epub 2016 Oct 24. Nucleic Acids Res. 2017. PMID: 28053165 Free PMC article.
-
MICheck: a web tool for fast checking of syntactic annotations of bacterial genomes.Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W471-9. doi: 10.1093/nar/gki498. Nucleic Acids Res. 2005. PMID: 15980515 Free PMC article.
-
[Development of antituberculous drugs: current status and future prospects].Kekkaku. 2006 Dec;81(12):753-74. Kekkaku. 2006. PMID: 17240921 Review. Japanese.
-
Comparative genomics of the mycobacteria.Int J Med Microbiol. 2000 May;290(2):143-52. doi: 10.1016/S1438-4221(00)80083-1. Int J Med Microbiol. 2000. PMID: 11045919 Review.
Cited by
-
Significant under expression of the DosR regulon in M. tuberculosis complex lineage 6 in sputum.Tuberculosis (Edinb). 2017 May;104:58-64. doi: 10.1016/j.tube.2017.03.001. Epub 2017 Mar 4. Tuberculosis (Edinb). 2017. PMID: 28454650 Free PMC article.
-
Transcriptional regulator-induced phenotype screen reveals drug potentiators in Mycobacterium tuberculosis.Nat Microbiol. 2021 Jan;6(1):44-50. doi: 10.1038/s41564-020-00810-x. Epub 2020 Nov 16. Nat Microbiol. 2021. PMID: 33199862 Free PMC article.
-
Decoding the similarities and differences among mycobacterial species.PLoS Negl Trop Dis. 2017 Aug 30;11(8):e0005883. doi: 10.1371/journal.pntd.0005883. eCollection 2017 Aug. PLoS Negl Trop Dis. 2017. PMID: 28854187 Free PMC article.
-
TIBLE: a web-based, freely accessible resource for small-molecule binding data for mycobacterial species.Database (Oxford). 2017 Jan 1;2017:bax041. doi: 10.1093/database/bax041. Database (Oxford). 2017. PMID: 29220433 Free PMC article.
-
Sputum is a surrogate for bronchoalveolar lavage for monitoring Mycobacterium tuberculosis transcriptional profiles in TB patients.Tuberculosis (Edinb). 2016 Sep;100:89-94. doi: 10.1016/j.tube.2016.07.004. Epub 2016 Jul 25. Tuberculosis (Edinb). 2016. PMID: 27553415 Free PMC article.
References
-
- World Health Organization . Global tuberculosis report. 2015.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

