Fast and efficient QTL mapper for thousands of molecular phenotypes
- PMID: 26708335
- PMCID: PMC4866519
- DOI: 10.1093/bioinformatics/btv722
Fast and efficient QTL mapper for thousands of molecular phenotypes
Abstract
Motivation: In order to discover quantitative trait loci, multi-dimensional genomic datasets combining DNA-seq and ChiP-/RNA-seq require methods that rapidly correlate tens of thousands of molecular phenotypes with millions of genetic variants while appropriately controlling for multiple testing.
Results: We have developed FastQTL, a method that implements a popular cis-QTL mapping strategy in a user- and cluster-friendly tool. FastQTL also proposes an efficient permutation procedure to control for multiple testing. The outcome of permutations is modeled using beta distributions trained from a few permutations and from which adjusted P-values can be estimated at any level of significance with little computational cost. The Geuvadis & GTEx pilot datasets can be now easily analyzed an order of magnitude faster than previous approaches.
Availability and implementation: Source code, binaries and comprehensive documentation of FastQTL are freely available to download at http://fastqtl.sourceforge.net/
Contact: emmanouil.dermitzakis@unige.ch or olivier.delaneau@unige.ch
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press.
Figures
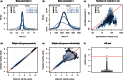
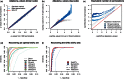
References
-
- Aulchenko Y.S. et al. (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics, 23, 1294–1296. - PubMed
-
- Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., 57, 289–300.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

