Additive roles of PthAs in bacterial growth and pathogenicity associated with nucleotide polymorphisms in effector-binding elements of citrus canker susceptibility genes
- PMID: 26709719
- PMCID: PMC6638360
- DOI: 10.1111/mpp.12359
Additive roles of PthAs in bacterial growth and pathogenicity associated with nucleotide polymorphisms in effector-binding elements of citrus canker susceptibility genes
Abstract
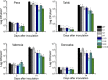
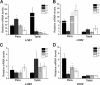
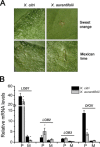
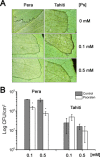
Citrus canker, caused by Xanthomonas citri, affects most commercial citrus varieties. All X. citri strains possess at least one transcription activator-like effector of the PthA family that activates host disease susceptibility (S) genes. The X. citri strain 306 encodes four PthA effectors; nevertheless, only PthA4 is known to elicit cankers on citrus. As none of the PthAs act as avirulence factors on citrus, we hypothesized that PthAs 1-3 might also contribute to pathogenicity on certain hosts. Here, we show that, although PthA4 is indispensable for canker formation in six Brazilian citrus varieties, PthAs 1 and 3 contribute to canker development in 'Pera' sweet orange, but not in 'Tahiti' lemon. Deletions in two or more pthA genes reduce bacterial growth in planta more pronouncedly than single deletions, suggesting an additive role of PthAs in pathogenicity and bacterial fitness. The contribution of PthAs 1 and 3 in canker formation in 'Pera' plants does not correlate with the activation of the canker S gene, LOB1 (LATERAL ORGAN BOUNDARIES 1), but with the induction of other PthA targets, including LOB2 and citrus dioxygenase (DIOX). LOB1, LOB2 and DIOX show differential PthA-dependent expression between 'Pera' and 'Tahiti' plants that appears to be associated with nucleotide polymorphisms found at or near PthA-binding sites. We also present evidence that LOB1 activation alone is not sufficient to elicit cankers on citrus, and that DIOX acts as a canker S gene in 'Pera', but not 'Tahiti', plants. Our results suggest that the activation of multiple S genes, such as LOB1 and DIOX, is necessary for full canker development.
Keywords: LATERAL ORGAN BOUNDARIES genes; TAL effectors; Xanthomonas aurantifolii; Xanthomonas citri; citrus canker; citrus dioxygenase; pthA.
© 2015 BSPP and John Wiley & Sons Ltd.
Figures




References
-
- Al‐Saadi, A. , Reddy, J. , Duan, Y. , Brunings, A. , Yuan, Q. and Gabriel, D. (2007) All five host‐range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host‐range variation. Mol. Plant–Microbe Interact. 20, 934–943. - PubMed
-
- Boch, J. and Bonas, U. (2010) Xanthomonas AvrBs3 family‐type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. - PubMed
-
- Boch, J. , Scholze, H. , Schornack, S. , Landgraf, A. , Hahn, S. , Kay, S. , Lahaye, T. , Nickstadt, A. and Bonas, U. (2009) Breaking the code of DNA binding specificity of TAL‐type III effectors. Science, 326, 1509–1512. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

