Identification of Thyroid Hormones and Functional Characterization of Thyroid Hormone Receptor in the Pacific Oyster Crassostrea gigas Provide Insight into Evolution of the Thyroid Hormone System
- PMID: 26710071
- PMCID: PMC4692385
- DOI: 10.1371/journal.pone.0144991
Identification of Thyroid Hormones and Functional Characterization of Thyroid Hormone Receptor in the Pacific Oyster Crassostrea gigas Provide Insight into Evolution of the Thyroid Hormone System
Abstract
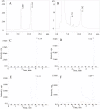
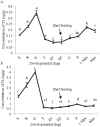
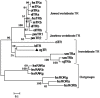
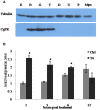
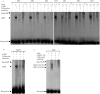
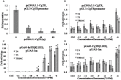
Thyroid hormones (THs) play important roles in development, metamorphosis, and metabolism in vertebrates. During the past century, TH functions were regarded as a synapomorphy of vertebrates. More recently, accumulating evidence has gradually convinced us that TH functions also occur in invertebrate chordates. To date, however, TH-related studies in non-chordate invertebrates have been limited. In this study, THs were qualitatively detected by two reliable methods (HPLC and LC/MS) in a well-studied molluscan species, the Pacific oyster Crassostrea gigas. Quantitative measurement of THs during the development of C. gigas showed high TH contents during embryogenesis and that oyster embryos may synthesize THs endogenously. As a first step in elucidating the TH signaling cascade, an ortholog of vertebrate TH receptor (TR), the most critical gene mediating TH effects, was cloned in C. gigas. The sequence of CgTR has conserved DNA-binding and ligand-binding domains that normally characterize these receptors. Experimental results demonstrated that CgTR can repress gene expression through binding to promoters of target genes and can interact with oyster retinoid X receptor. Moreover, CgTR mRNA expression was activated by T4 and the transcriptional activity of CgTR promoter was repressed by unliganded CgTR protein. An atypical thyroid hormone response element (CgDR5) was found in the promoter of CgTR, which was verified by electrophoretic mobility shift assay (EMSA). These results indicated that some of the CgTR function is conserved. However, the EMSA assay showed that DNA binding specificity of CgTR was different from that of the vertebrate TR and experiments with two dual-luciferase reporter systems indicated that l-thyroxine, 3,3',5-triiodothyronine, and triiodothyroacetic acid failed to activate the transcriptional activity of CgTR. This is the first study to functionally characterize TR in mollusks. The presence of THs and the functions of CgTR in mollusks contribute to better understanding of the evolution of the TH system.
Conflict of interest statement
Figures

References
-
- Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81(3):1097–142. ISI:000169570800005. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

