Exercise Capacity and Response to Training Quantitative Trait Loci in a NZW X 129S1 Intercross and Combined Cross Analysis of Inbred Mouse Strains
- PMID: 26710100
- PMCID: PMC4692404
- DOI: 10.1371/journal.pone.0145741
Exercise Capacity and Response to Training Quantitative Trait Loci in a NZW X 129S1 Intercross and Combined Cross Analysis of Inbred Mouse Strains
Abstract
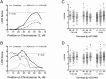
Genetic factors determining exercise capacity and the magnitude of the response to exercise training are poorly understood. The aim of this study was to identify quantitative trait loci (QTL) associated with exercise training in mice. Based on marked differences in training responses in inbred NZW (-0.65 ± 1.73 min) and 129S1 (6.18 ± 3.81 min) mice, a reciprocal intercross breeding scheme was used to generate 285 F2 mice. All F2 mice completed an exercise performance test before and after a 4-week treadmill running program, resulting in an increase in exercise capacity of 1.54 ± 3.69 min (range = -10 to +12 min). Genome-wide linkage scans were performed for pre-training, post-training, and change in run time. For pre-training exercise time, suggestive QTL were identified on Chromosomes 5 (57.4 cM, 2.5 LOD) and 6 (47.8 cM, 2.9 LOD). A significant QTL for post-training exercise capacity was identified on Chromosome 5 (43.4 cM, 4.1 LOD) and a suggestive QTL on Chromosomes 1 (55.7 cM, 2.3 LOD) and 8 (66.1 cM, 2.2 LOD). A suggestive QTL for the change in run time was identified on Chromosome 6 (37.8 cM, 2.7 LOD). To identify shared QTL, this data set was combined with data from a previous F2 cross between B6 and FVB strains. In the combined cross analysis, significant novel QTL for pre-training exercise time and change in exercise time were identified on Chromosome 12 (54.0 cM, 3.6 LOD) and Chromosome 6 (28.0 cM, 3.7 LOD), respectively. Collectively, these data suggest that combined cross analysis can be used to identify novel QTL and narrow the confidence interval of QTL for exercise capacity and responses to training. Furthermore, these data support the use of larger and more diverse mapping populations to identify the genetic basis for exercise capacity and responses to training.
Conflict of interest statement
Figures




References
-
- Blair SN, Kohl HW 3rd, Barlow CE, Paffenbarger RS Jr., Gibbons LW, Macera CA. Changes in physical fitness and all-cause mortality. A prospective study of healthy and unhealthy men. JAMA. 1995;273(14):1093–8. . - PubMed
-
- Erikssen G, Liestol K, Bjornholt J, Thaulow E, Sandvik L, Erikssen J. Changes in physical fitness and changes in mortality. Lancet. 1998;352(9130):759–62. . - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

