Insight into the Human DNA Primase Interaction with Template-Primer
- PMID: 26710848
- PMCID: PMC4813500
- DOI: 10.1074/jbc.M115.704064
Insight into the Human DNA Primase Interaction with Template-Primer
Abstract
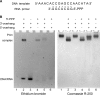
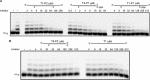
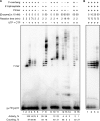
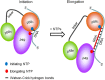
DNA replication in almost all organisms depends on the activity of DNA primase, a DNA-dependent RNA polymerase that synthesizes short RNA primers of defined size for DNA polymerases. Eukaryotic and archaeal primases are heterodimers consisting of small catalytic and large accessory subunits, both of which are necessary for the activity. The mode of interaction of primase subunits with substrates during the various steps of primer synthesis that results in the counting of primer length is not clear. Here we show that the C-terminal domain of the large subunit (p58C) plays a major role in template-primer binding and also defines the elements of the DNA template and the RNA primer that interact with p58C. The specific mode of interaction with a template-primer involving the terminal 5'-triphosphate of RNA and the 3'-overhang of DNA results in a stable complex between p58C and the DNA/RNA duplex. Our results explain how p58C participates in RNA synthesis and primer length counting and also indicate that the binding site for initiating NTP is located on p58C. These findings provide notable insight into the mechanism of primase function and are applicable for DNA primases from other species.
Keywords: 5′-triphosphate; DNA primase; DNA replication; DNA-protein interaction; RNA primer length counting; RNA synthesis; RNA/DNA duplex; gel electrophoresis; human; p58 subunit; primase activity.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Loeb L. A., and Monnat R. J. Jr. (2008) DNA polymerases and human disease. Nat. Rev. Genet. 9, 594–604 - PubMed
-
- Garg P., and Burgers P. M. (2005) DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40, 115–128 - PubMed
-
- Pellegrini L. (2012) The Polα-primase complex. Subcell. Biochem. 62, 157–169 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

